DCM-C
BGUi008-A
General
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Bedouin |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Disease associated phenotypes | no phenotypes |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
Yes
46 XY
Karyotyping method:
G-Banding
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA8152178 |
Ethics
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | Yes |
| Was the consent voluntarily given? | Yes |
| Has the donor been informed that participation will not directly influence their personal treatment? | Yes |
| Can you provide us with a copy of the Donor Information Sheet provided to the donor? | No |
| Do you (Depositor/Provider) hold the original Donor Consent Form? | No |
| If you do not hold the Donor Consent Form, do you know who does? | Yes |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | anonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | Yes |
| Does consent prevent CELLS DERIVED FROM THE DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
| How may genetic information associated with the cell line be accessed? | No information |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the PROPOSED PROJECT, involving use of donated embryo/tissue or derived cells? | No |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? |
hIPSC Derivation
General |
|
| Source cell type |
Dermal fibroblasts are the major cell type in dermis and are commonly accepted as terminally differentiated cells.
|
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Is reprogramming vector detectable? |
No |
| Methods used |
PCR
|
Vector free reprogramming |
|
Other |
|
| Derived under xeno-free conditions |
Unknown |
| Derived under GMP? |
Unknown |
| Available as clinical grade? |
Unknown |
Culture Conditions
| Medium |
Other medium:
Base medium: NutriStem® hPSC XF
Main protein source: Serum concentration: % |
Characterisation
Analysis of Undifferentiated Cells
Differentiation Potency
In vitro spontaneous differentiation
| Marker | Expressed |
| AFP |
Yes |
In vitro spontaneous differentiation
| Marker | Expressed |
| ACTA2 |
Yes |
Morphology
In vitro spontaneous differentiation
| Marker | Expressed |
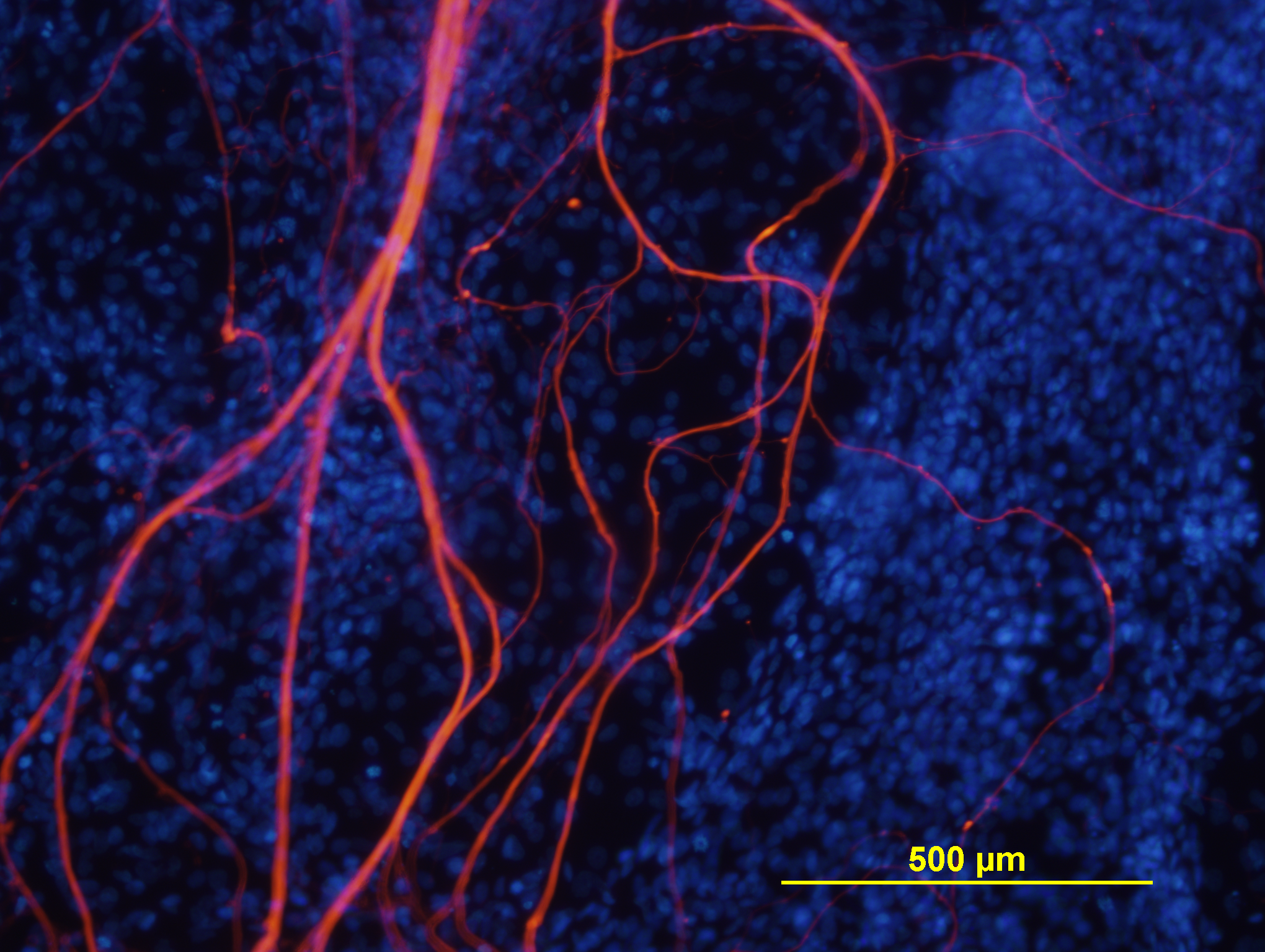
| NEFL |
Unknown |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|



Login to share your feedback, experiences or results with the research community.