NY0003-01-ES-001
NYSCFe003-A
General
Cell Line |
|
| hPSCreg name | NYSCFe003-A |
| Cite as: | NYSCFe003-A (RRID:CVCL_VE10) |
| Alternative name(s) |
NY0003-01-ES-001
|
| Cell line type | Human embryonic stem cell (hESC) |
| Similar lines | No similar lines found. |
| Last update | 24th June 2021 |
| User feedback | |
Provider |
|
| Generator | New York Stem Cell Foundation Research Institute (NYSCF) |
| Owner | New York Stem Cell Foundation Research Institute (NYSCF) |
| Distributors | |
| Derivation country | United States |
External Databases |
|
| BioSamples | SAMEA104617259 |
| Cellosaurus | CVCL_VE10 |
| Wikidata | Q54931471 |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Clinical use: not allowed
Commercial use: allowed
|
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | White |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Disease associated phenotypes | no phenotypes |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
No
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA104617260 |
Ethics
| Was the embryo established purely for research purposes? | No |
| Have both parents consented to the use of the embryo for ESC derivation? | Yes |
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | Yes |
| Was the consent voluntarily given? | Yes |
| Alternatives to consent are available? | Yes |
| Alternatives to consent | |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | pseudonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | Yes |
| * Does consent expressly prevent the derivation of pluripotent stem cells? | No |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | Western Institutional Review Board |
| Approval number | Protocol # 20101912 |
hESC Derivation
| Date of derivation | 2016-04-28 |
| Embryo stage | Blastula with ICM and Trophoblast |
| Supernumerary embryos from IVF treatment? |
Yes
|
| PGD Embryo? |
No |
| Expansion status |
3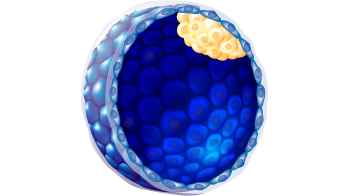
|
| ICM morphology |
C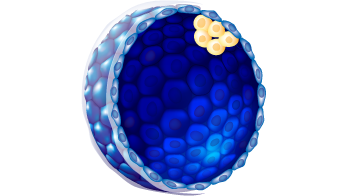
|
| Trophectoderm morphology |
g
|
| Cell isolation | Laser |
| Cell seeding | Isolated ICM |
| Derived under xeno-free conditions? |
No |
| Derivation under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzymatically
TrypLE
|
| O2 Concentration | 20 % |
| CO2 Concentration | 5 % |
| Medium |
Other medium:
Base medium: DMEM/F12+ Glutamax (1X)
Main protein source: Albumine Serum concentration: % |
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|
||||
| SOX2 |
Yes |
|
||||
| NANOG |
Yes |
|
||||
| TRA 1-81 |
Yes |
|||||
| TRA 1-60 |
Yes |
|||||
| SSEA-4 |
Yes |
Score:
| Marker | Present | Absent |
| mCpG | ||
| OCT4 |
Self-renewal
Negative
Endoderm
Negative
Mesoderm
Negative
Ectoderm score
Negative
Differentiation Potency
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Certificate of Analysis |
|
| Is there a certificate of analysis available? |
Yes
Passage:
8
|
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
SNP typing array
|


Login to share your feedback, experiences or results with the research community.