LVIP04-LC12-1-BE1, VS-CTS-RD3-BE1, Clone 27
LVPEIi006-B-1
General
Cell Line |
|
| hPSCreg name | LVPEIi006-B-1 |
| Cite as: | LVPEIi006-B-1 |
| Alternative name(s) |
LVIP04-LC12-1-BE1, VS-CTS-RD3-BE1, Clone 27
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
|
| Last update | 18th September 2025 |
| Notes | This is an heterozygous, mutation-corrected, isogenic iPSC line (LVPEIi006-B-1), with mutation corrected in one allele of RD3 and a parental mutant allele. This line was generated using an engineered en31FnCas9-based adenine base editor system. This isogenic line was clonally expanded beyond passage 20 and genotyped to confirm the desired base correction. It stably maintained its stemness, pluripotency, genomic integrity and could differentiate into retinal organoids. The correction of pathogenic mutation in one of the alleles has resulted in the partial restoration of normal RD3 mRNA splicing in mature retinal organoids. |
| User feedback | |
Provider |
|
| Generator |
L.V. Prasad Eye Institute (LVPEI)
Contact:
L.V. Prasad Eye Institute (LVPEI) |
| Owner | L.V. Prasad Eye Institute (LVPEI) |
| Distributors | |
| Derivation country | India |
External Databases |
|
| BioSamples | SAMEA117688009 |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Asian, Indian (27 years) |
Phenotype and Disease related information (Donor) |
|
| Diseases | A disease was diagnosed.
|
| Is the medical history available upon request? | Yes |
| Is clinical information available? | Yes |
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA115158754 |
Ethics
Also have a look at the ethics information for the parental line
LVPEIi006-B
.
| Is there an MTA available for the cell line? | Yes |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? | The gene editing construct, en31FnCas9-ABEmax8.17d, was obtained from the collaborator Dr. Debojyoti Chakraborty's lab at CSIR-IGIB, New Delhi, India. The RD3 mutation-specific CRISPR guide oligo was then cloned into the BbsI site, downstream of the U6 promoter. |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | Yes |
| Constraints for use or distribution | For research use only |
hIPSC Derivation
General |
|
|
The source cell information can be found in the parental cell line
LVPEIi006-B.
|
|
| Passage number reprogrammed | NA |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Genes | |
| Is reprogramming vector detectable? |
No |
| Methods used |
RT-PCR
|
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | Clones were seeded in low density and clonally expanded to get edited clone |
| Derived under xeno-free conditions |
Yes |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Vitronectin |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
EDTA
|
| CO2 Concentration | 5 % |
| Medium | Essential 8™ |
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
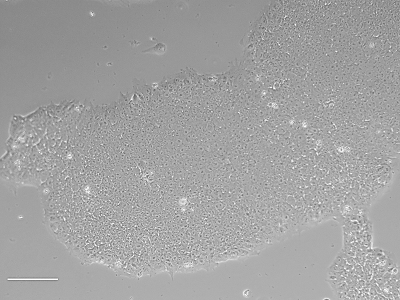
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| SOX2 |
Yes |
|
|
|
||
| KLF4 |
Yes |
|
|
|||
| NANOG |
Yes |
|
|
|||
| hTERT |
Yes |
|
||||
| SSEA-4 |
Yes |
|
Differentiation Potency
In vitro spontaneous differentiation
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Disease/phenotype related modifications |
|


Login to share your feedback, experiences or results with the research community.