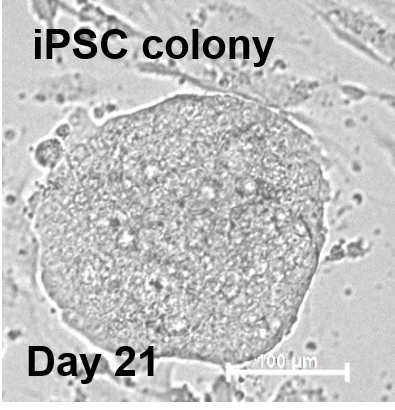
hiPSC-FS.2
UKJi005-A
General
Cell Line |
|
| hPSCreg name | UKJi005-A |
| Cite as: | UKJi005-A (RRID:CVCL_E7UT) |
| Alternative name(s) |
hiPSC-FS.2
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines | No similar lines found. |
| Last update | 25th June 2024 |
| Notes | The hiPSC-FS.2 was generated from the Human Foreskin; The cell line was established by ATCC in 2003 from normal human foreskin(HFF-1-SCRC-1041). Reprogramming HFF-1 cell with the CytoTune®-iPS Sendai Reprogramming Kit (Life Technologies A1378001). |
| User feedback | |
Provider |
|
| Generator | Universitätsklinikum Jena (UKJ), Klinik für Innere Medizin I (KIM I), Dr. M. Bekhite ELsaied (UKJ) |
| Owner | Universitätsklinikum Jena (UKJ), Klinik für Innere Medizin I (KIM I), Dr. M. Bekhite ELsaied (UKJ) |
| Distributors | |
| Derivation country | Germany |
External Databases |
|
| BioSamples | SAMEA115715116 |
| Cellosaurus | CVCL_E7UT |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: not allowed
Additional restrictions:
Permitted the hiPSC-FS.2 to be used exclusively for research purposes following the signing of a collaboration agreement. |
Donor Information
General Donor Information |
|
| Sex | male |
| Age of donor (at collection) | neonate |
| Ethnicity | Age: Neonate - Ethnicity: N/A |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Disease associated phenotypes | no phenotypes |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
Unknown
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA115715117 |
Ethics
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | No |
| Was the consent voluntarily given? | No |
| Has the donor been informed that participation will not directly influence their personal treatment? | No |
| Can you provide us with a copy of the Donor Information Sheet provided to the donor? | No |
| Do you (Depositor/Provider) hold the original Donor Consent Form? | No |
| If you do not hold the Donor Consent Form, do you know who does? | Yes |
| Alternatives to consent are available? | Yes |
| Alternatives to consent | Ethical approvement 2020-1833_1-Material form University of Jena Ethics Committee indicating permission to use the biological samples for to generate induced pluripotent stem cells to study cardiomyocytes |
| Alternative consent approval number | |
| Has the donor agreed to be re-contacted? | Unknown |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | anonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | Yes |
| Does consent expressly prevent development of commercial products? | No |
| Does consent expressly prevent financial gain from any use of the donated embryo/tissue, including any product made from it? | No |
| Does consent expressly permit storage of donated embryo/tissue for an unlimited time? | Yes |
| Does consent expressly permit storage of cells derived from the donated embryo/tissue for an unlimited time? | Yes |
| Does consent prevent the DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
| Does consent prevent CELLS DERIVED FROM THE DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
Does consent permit research by | |
| an academic institution? | Yes |
| a public organisation? | No |
| a non-profit company? | No |
| a for-profit corporation? | No |
| How may genetic information associated with the cell line be accessed? | Controlled Access |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Does the consent anticipate that the donor will be notified of results or outcomes of any research involving the donated samples or derived cells? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | University of Jena Ethics Committee |
| Approval number | 2020-1833_1-Material |
| Do you have obligations to third parties in regard to the use of the cell line? | No |
| Are you aware of any further constraints on the use of the donated embryo/tissue or derived cells? | No |
| Is there an MTA available for the cell line? | No |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? | Invitrogen, Thermo Fisher Scientific |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hIPSC Derivation
General |
|
| Source cell line name |
HFF-1 (ATCC SCRC-1041) Derived from same source line (potentially other lot and donor, see below):
|
| Source cell type |
disease: feeder layer
|
| Source cell origin | |
| Source cell type (free text) | human foreskin pooled from two individuals |
| Age of donor (at collection) | neonate |
| Collected in | 2003 |
| Source cell line vendor | ATCC |
| Passage number reprogrammed | 8 |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Genes | |
| Is reprogramming vector detectable? |
No |
| Methods used |
PCR
|
| Files and images showing reprogramming vector expressed or silenced | |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | IPSC clones were manually picked and cultured on Matrigel (Corning) coated plate using mTeSR1 (Stem Cell) at 37 °C, 5% CO 2. We pick a single colony from passage 1 and subcloning for 10 passages and then testing for virus free iPSC. |
| Derived under xeno-free conditions |
Yes |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
ReLeSR™ is an enzyme-free reagent for dissociation and passaging of human embryonic stem (ES) or induced pluripotent stem (iPS) cells
|
| O2 Concentration | 20 % |
| CO2 Concentration | 5 % |
| Medium | Essential 8™ |
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| TRA 1-60 |
Yes |
|||||
| SOX2 |
Yes |
|||||
| SSEA-4 |
Yes |
Embryoid body (EB) differentiation
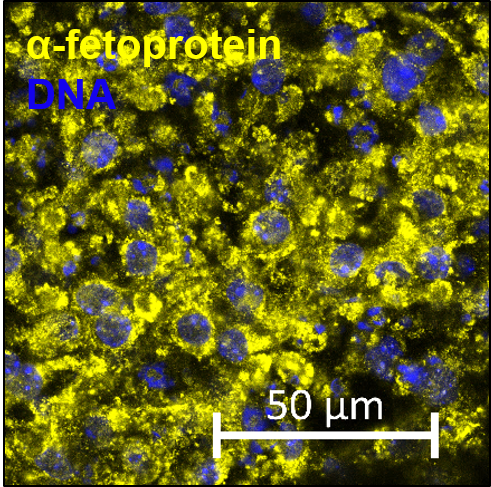
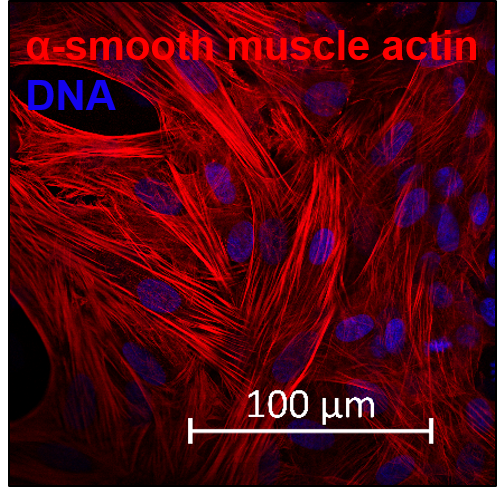
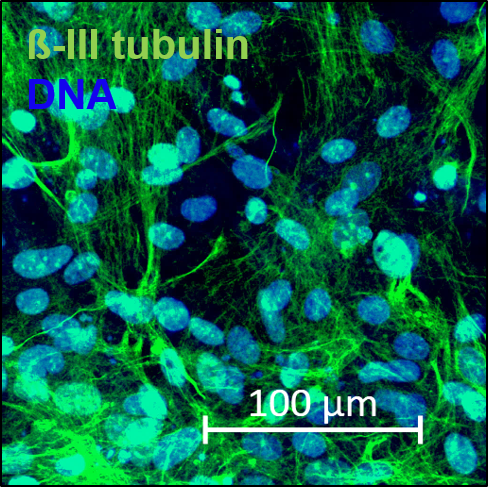
The confluent culture of hiPSC-FS.2 was dissociated using a ReLeSR dissociation reagent. EBs were generated by aggregating the cells, using ultra-low attachment plates, in an E8 medium containing 10 µm ROCK inhibitor Y-27632 (Santa Cruz Biotechnologies) for two days on an orbital shaker. Cell aggregates were then further cultured in DMEM containing 20 % fetal bovine serum, 2 mM L-glutamine, 55 µM ß-mercaptoethanol, and 1x non-essential amino acids. The differentiation medium was changed every other day for 3 months. Spontaneous differentiated EBs were fixed at 4 % PFA and processed according to standard procedures for paraffin embedding and hematoxylin/eosin staining.
Method documentation
Differentiation Potency
In vitro spontaneous differentiation
| Marker | Expressed |
| TUBB3 tubulin beta 3 class III [ Homo sapiens (human) ] |
Yes |
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
Karyotyping shows the genomic integrity in iPSC after reprogramming
Passage number: 15
Karyotyping method:
G-Banding
|
Other Genotyping (Cell Line) |
|


Login to share your feedback, experiences or results with the research community.