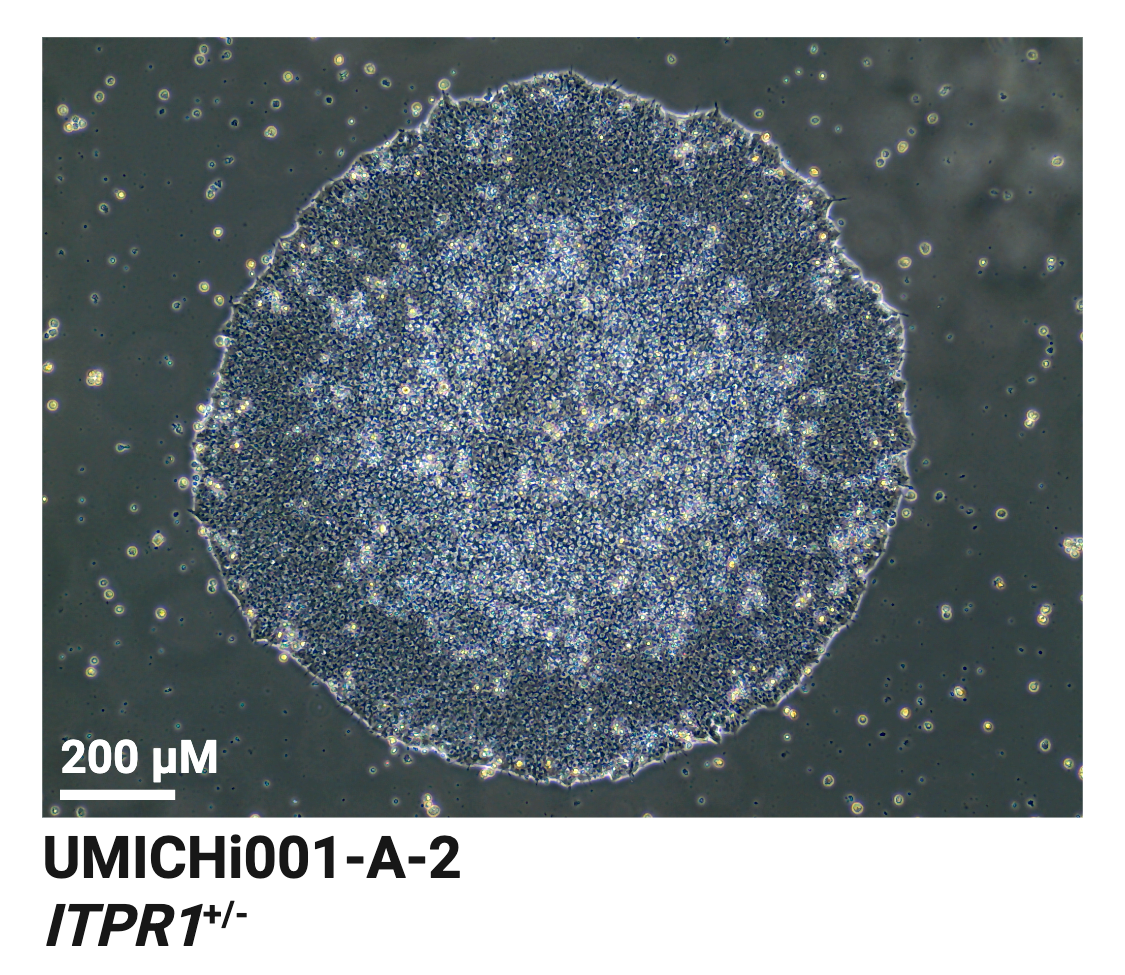
ITPR1 Het, ITPR1+/-
The cell line is not validated yet.
UMICHi001-A-2
General
Cell Line |
|
| hPSCreg name | UMICHi001-A-2 |
| Cite as: | UMICHi001-A-2 |
| Alternative name(s) |
ITPR1 Het, ITPR1+/-
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines | No similar lines found. |
| Last update | 20th October 2025 |
| Notes | Internal experimental control was generated by undergoing the same experimental procedure and is registered as UMICHi001-A-1. Contact information: Dr. Hayley McLoughlin (hayleymc@umich.edu) |
| User feedback | |
Provider |
|
| Generator |
University of Michigan (UMICH)
Contact:
University of Michigan (UMICH) |
| Owner | University of Michigan (UMICH) |
| Distributors | |
| Derivation country | United States |
External Databases |
|
General Information |
|
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: allowed
Commercial use: allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Caucasian |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMN02324437 |
Ethics
Also have a look at the ethics information for the parental line
UMICHi001-A
.
| Is there an MTA available for the cell line? | No |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? | |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hIPSC Derivation
General |
|
|
The source cell information can be found in the parental cell line
UMICHi001-A.
|
|
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Genes | |
| Is reprogramming vector detectable? |
Unknown |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | Clones were reprogrammed and selected by Edit Co. (Redwood City, CA, USA) based on a positive PluriTest and immunohistochemical analysis |
| Derived under xeno-free conditions |
Unknown |
| Derived under GMP? |
Unknown |
| Available as clinical grade? |
Unknown |
Culture Conditions
| Surface coating | iMatrix-511 |
| Feeder cells |
No |
| Passage method |
Enzymatically
Accutase
|
| O2 Concentration | 21 % |
| CO2 Concentration | 5 % |
| Medium |
mTeSR™ Plus
|
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| NANOG |
Yes |
|||||
| SOX2 |
Yes |
Differentiation Potency
In vitro directed differentiation
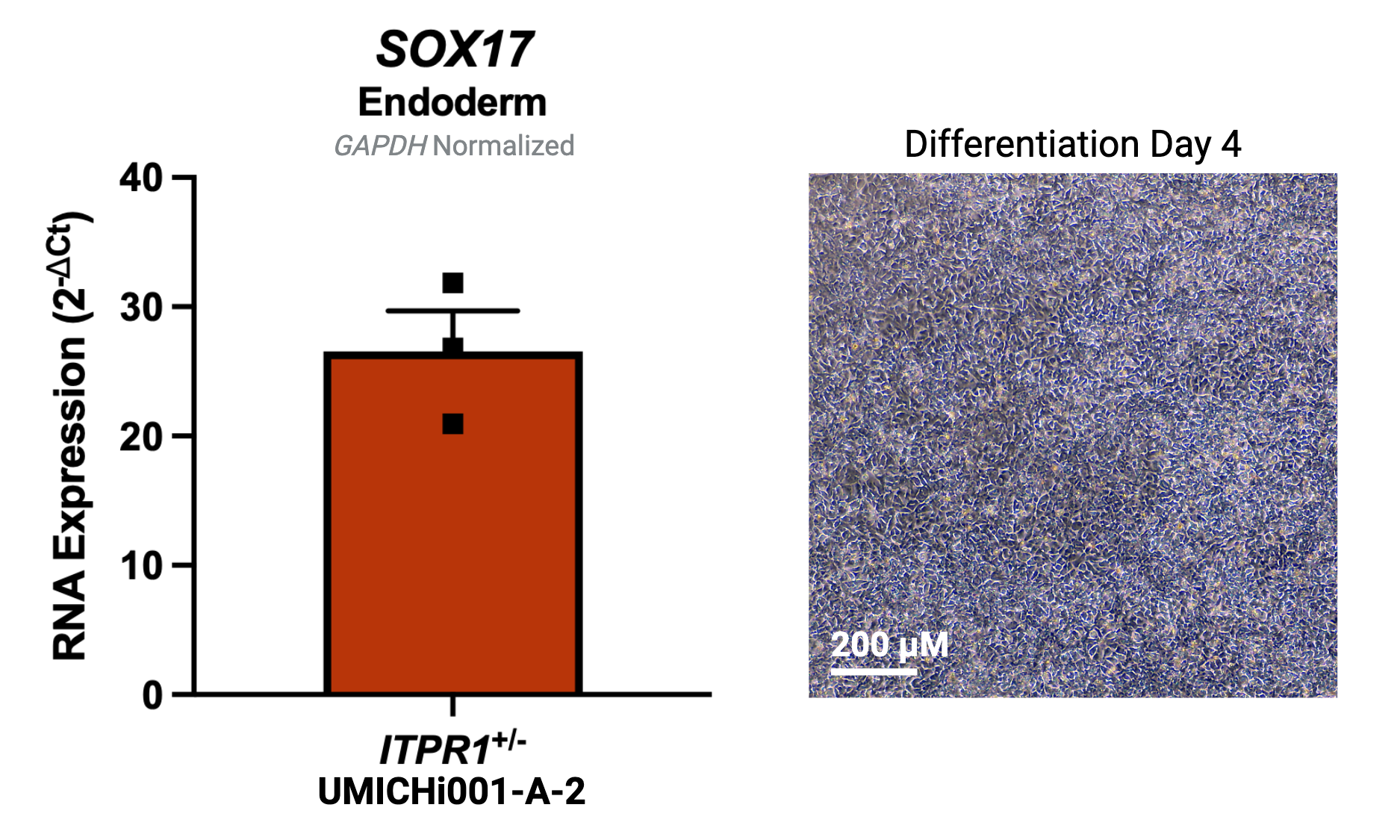
| Marker | Expressed |
| SOX17 |
Yes |
Protocol or reference
In vitro directed differentiation
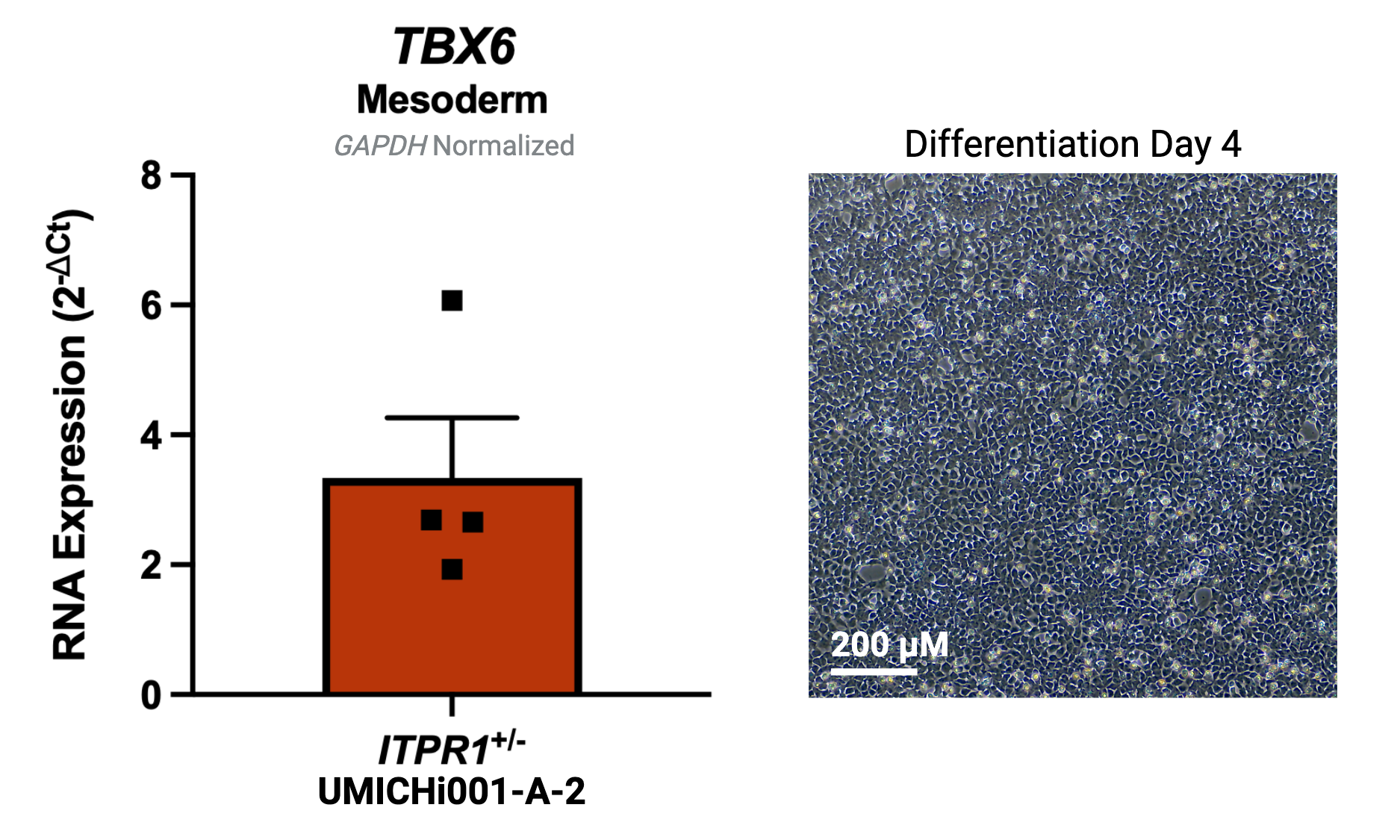
| Marker | Expressed |
| TBX6 |
Yes |
Protocol or reference
In vitro directed differentiation
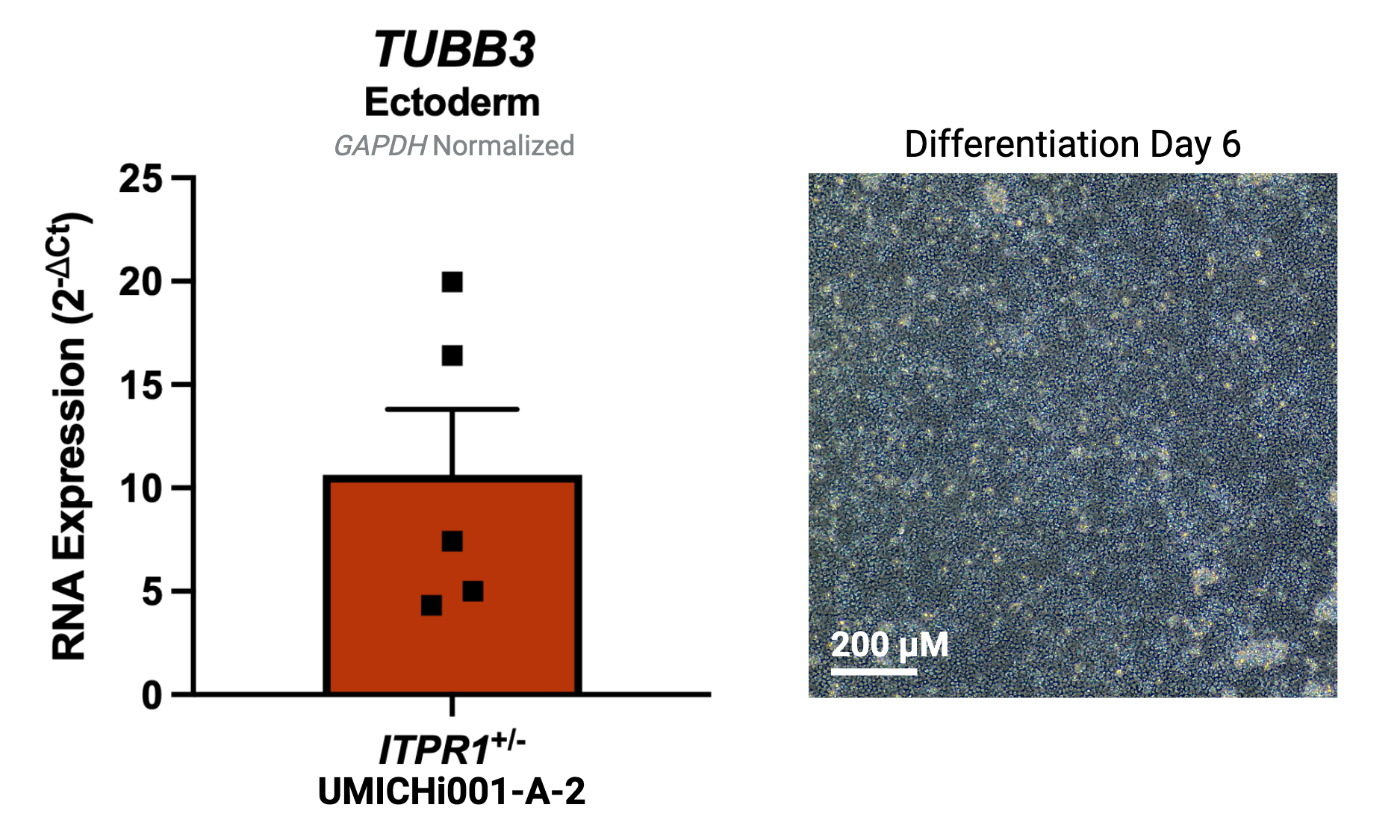
| Marker | Expressed |
| TUBB3 |
Yes |
Protocol or reference
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
46,XY
Passage number: 7
Karyotyping method:
G-Banding
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Disease/phenotype related modifications |
|


Login to share your feedback, experiences or results with the research community.