CL03
UPSFRe003-A
General
Cell Line |
|
| hPSCreg name | UPSFRe003-A |
| Cite as: | UPSFRe003-A (RRID:CVCL_C798) |
| Alternative name(s) |
CL03
|
| Cell line type | Human embryonic stem cell (hESC) |
| Similar lines |
|
| Last update | 30th August 2022 |
| User feedback | |
Provider |
|
| Generator |
Université Paris-Sud 11 (UPSFR)
Contact:
ESTeam Paris Sud |
| Derivation country | France |
External Databases |
|
| Cellosaurus | CVCL_C798 |
| Wikidata | Q54813495 |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes |
Donor Information
General Donor Information |
|
| Sex | male |
Phenotype and Disease related information (Donor) |
|
| Diseases | A disease was diagnosed.
|
| Disease associated phenotypes |
|
Ethics
| Alternatives to consent are available? | Yes |
| Alternatives to consent | |
| Alternative consent approval number | |
| How may genetic information associated with the cell line be accessed? | |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | Agence de la biomédecine |
| Approval number |
hESC Derivation
| Date of derivation | 2009-07-21 |
| Embryo stage | Blastula with ICM and Trophoblast |
| Supernumerary embryos from IVF treatment? |
Yes
|
| PGD Embryo? |
Yes |
| Expansion status |
6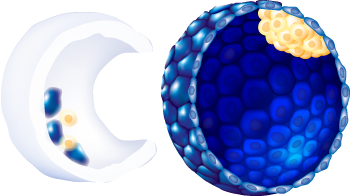
|
| ICM morphology |
A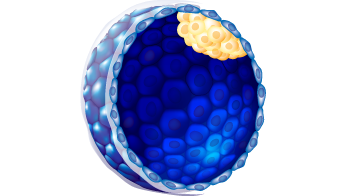
|
| Trophectoderm morphology |
a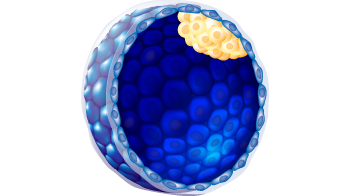
|
| ZP removal technique | Spontaneous Hatching |
| Cell isolation | None |
| Cell seeding | Embryo |
Culture Conditions
| Feeder cells |
Mouse Embryonic Fibroblast Cellfinder Ont Id: EFO_0004040 |
|||||||||
| CO2 Concentration | 5 % | |||||||||
| Medium |
Other medium:
Base medium: DMEM/F-12
Main protein source: Knock-out serum replacement Serum concentration: 20 % Supplements
|
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| Alkaline Phosphatase |
Yes |
|
||||
| DNMT3B |
Yes |
|
||||
| GDF3 |
Yes |
|
||||
| ZFP42 (REX-1) |
Yes |
|
||||
| CD9 |
Yes |
|
||||
| GABRB3 |
Yes |
|
||||
| LEFTB |
Yes |
|
||||
| LIN28 |
Yes |
|
||||
| NANOG |
Yes |
|
||||
| POU5F1 (OCT-4) |
Yes |
|
|
|||
| SOX2 |
Yes |
|
||||
| TDGF1 |
Yes |
|
||||
| SSEA-4 |
Yes |
|
||||
| TRA 1-60 |
Yes |
|
||||
| HESCA-1 |
Yes |
|
Differentiation Potency
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
46,XY,t(8;9)(p23;p24).ish t(8;9)(8pter-,9pter+,8qter+;9pter-,8pter+,9qter+)
Passage number: 13
|
Other Genotyping (Cell Line) |
|


Login to share your feedback, experiences or results with the research community.