iTaf9-11
ICGi016-A
General
Cell Line |
|
| hPSCreg name | ICGi016-A |
| Cite as: | ICGi016-A (RRID:CVCL_UF64) |
| Alternative name(s) |
iTaf9-11
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
LUMCi027-A (LUMC0153iPKP03) Donor's gene variants: PKP2 Donor diseases: arrhythmogenic right ventricular dysplasia 9 ATLABi001-A (AT-MODY3-iP001) Donor diseases: Hepatocyte Nuclear Factor 1-Alpha-Associated Monogenic Diabetes TAUi006-A (UTA.00102.LQT1) Donor's gene variants: KCNQ1 – potassium voltage-gated channel subfamily Q member 1 Donor diseases: Long QT Syndrome 1 TAUi006-B (UTA.00118.LQT1) Donor's gene variants: KCNQ1 – potassium voltage-gated channel subfamily Q member 1 Donor diseases: Long QT Syndrome 1 TAUi007-A (UTA.00208.LQT1) Donor's gene variants: KCNQ1 – potassium voltage-gated channel subfamily Q member 1 Donor diseases: Long QT Syndrome 1 TAUi007-B (UTA.00211.LQT1) Donor's gene variants: KCNQ1 – potassium voltage-gated channel subfamily Q member 1 Donor diseases: Long QT Syndrome 1 ATLABi002-A (AT-MODY3-iP002) Donor diseases: Hepatocyte Nuclear Factor 1-Alpha-Associated Monogenic Diabetes |
| Last update | 18th October 2019 |
| User feedback | |
Provider |
|
| Generator | Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences (ICG) |
| Distributors | |
| Derivation country | Russia |
External Databases |
|
| BioSamples | SAMEA5230331 |
| Cellosaurus | CVCL_UF64 |
| Wikidata | Q94313458 |
General Information |
|
| Publications |
|
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: not allowed
|
Donor Information
General Donor Information |
|
| Sex | female |
| Ethnicity | Caucasian |
Phenotype and Disease related information (Donor) |
|
| Diseases | A disease was diagnosed.
|
| Family history | de novo |
| Is the medical history available upon request? | yes, Research Institute of Medical Genetics, TNMRC |
| Is clinical information available? | yes, Research Institute of Medical Genetics, TNMRC |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
Yes
arr[GRCh38] 2p25.3(42444_2684871)x1 dn, 2p25.3-p23.3(2771354_24258056)x3 dn
Karyotyping method:
Array CGH
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA5230332 |
Ethics
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | Yes |
| Was the consent voluntarily given? | Yes |
| Has the donor been informed that participation will not directly influence their personal treatment? | Yes |
| Can you provide us with a copy of the Donor Information Sheet provided to the donor? | Yes |
| Do you (Depositor/Provider) hold the original Donor Consent Form? | Yes |
| Has the donor agreed to be re-contacted? | Unknown |
| Has the donor been informed about how her/his data will be protected? | Yes |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | pseudonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | Yes |
| Does consent prevent CELLS DERIVED FROM THE DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
| How may genetic information associated with the cell line be accessed? | Controlled Access |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | The study was approved by the Scientific Ethics Committee of Research Institute of Medical Genetics, Tomsk NRMC |
| Approval number | protocol number 106/2017 |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the PROPOSED PROJECT, involving use of donated embryo/tissue or derived cells? | Yes |
| Name of accrediting authority involved? | The study was approved by the Scientific Ethics Committee of Research Institute of Medical Genetics, Tomsk NRMC |
| Approval number | protocol number 106/2017 |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? |
hIPSC Derivation
General |
|
| Source cell type | |
| Passage number reprogrammed | 5 |
Reprogramming method |
|
| Vector type | Integrating |
| Vector | Virus (Lentivirus) |
| Genes | |
| Is the used vector excisable? |
No |
| Absence of reprogramming vector(s)? |
No |
| Reprogramming vectors silenced? |
Unknown |
| Vector map | |
Vector free reprogramming |
|
Other |
|
| Selection criteria for clones | morphology marker expression |
| Derived under xeno-free conditions |
No |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Gelatin | ||||||
| Feeder cells |
mouse embryonic fibroblasts Cellfinder Ont Id: EFO_0004040 |
||||||
| Passage method | Mechanically | ||||||
| CO2 Concentration | 5 % | ||||||
| Medium |
Other medium:
Base medium: DMEM/F12
Main protein source: Knock-out serum replacement Serum concentration: 20 % Supplements
|
||||||
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
||||||
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | Yes |
||||||
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| NANOG |
Yes |
|
||||
| POU5F1 (OCT-4) |
Yes |
|
||||
| TRA 1-60 |
Yes |
|
||||
| SSEA-3 |
Yes |
|
Score:
| Marker | Present | Absent |
| mCpG | ||
| OCT4 |
Differentiation Potency
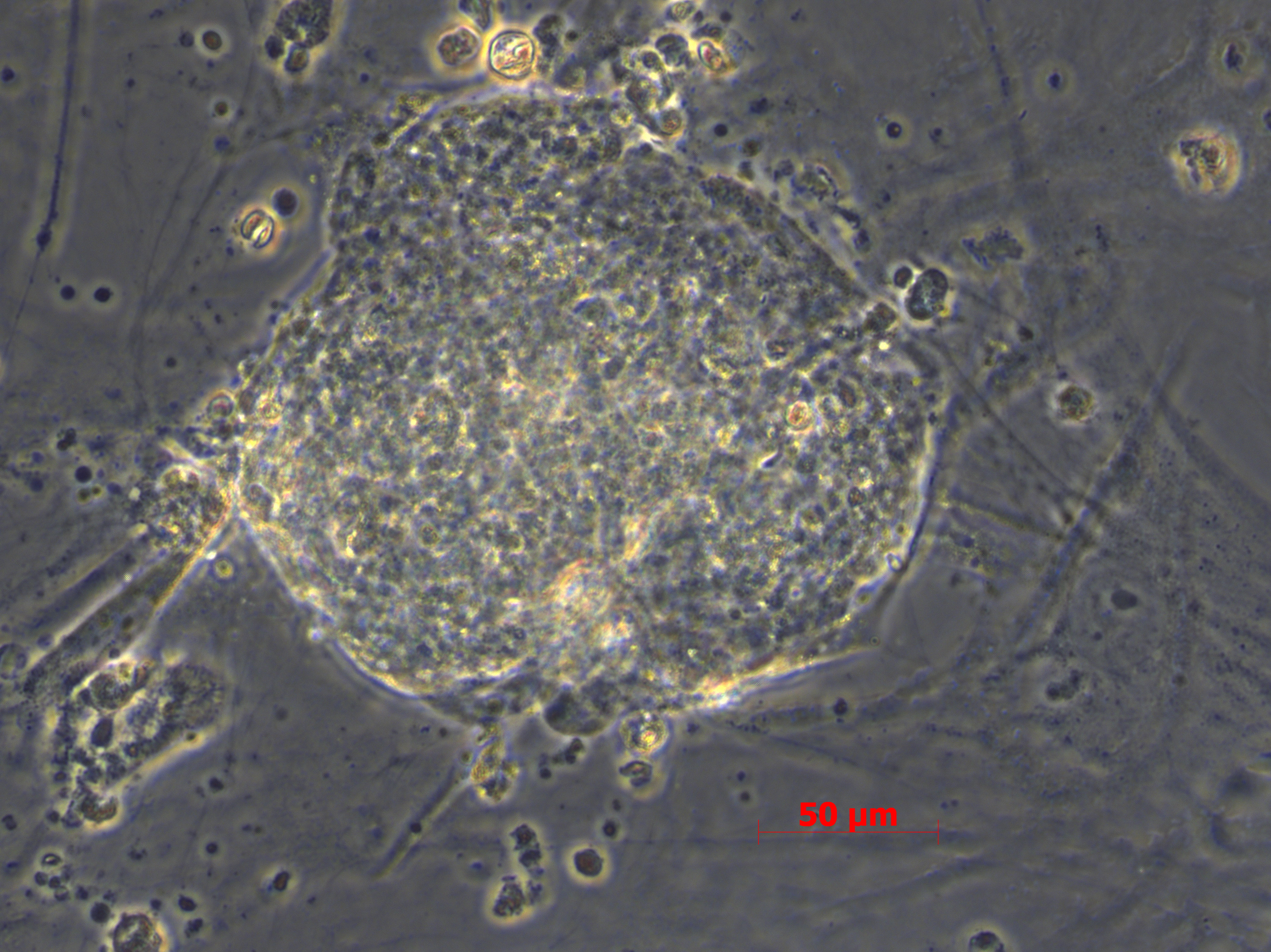
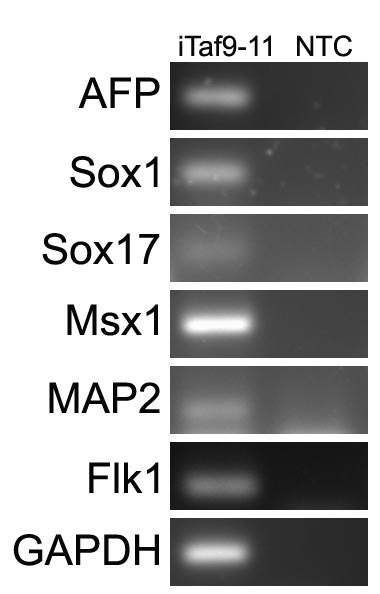
In vitro spontaneous differentiation
In vitro spontaneous differentiation
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|


Login to share your feedback, experiences or results with the research community.