Man-13
UMANe002-A
General
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Caucasian |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Disease associated phenotypes | no phenotypes |
| Is the medical history available upon request? | Yes |
| Is clinical information available? | Yes |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
Yes
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA7791418 |
Ethics
| Was the embryo established purely for research purposes? | No |
| Have both parents consented to the use of the embryo for ESC derivation? | Yes |
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | Yes |
| Was the consent voluntarily given? | Yes |
| Has the donor been informed that participation will not directly influence their personal treatment? | Yes |
| Can you provide us with a copy of the Donor Information Sheet provided to the donor? | No |
| Do you (Depositor/Provider) hold the original Donor Consent Form? | No |
| If you do not hold the Donor Consent Form, do you know who does? | Yes |
| Alternatives to consent are available? | Yes |
| Alternatives to consent | |
| Alternative consent approval number | |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | pseudonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | Yes |
| * Does consent expressly prevent the derivation of pluripotent stem cells? | No |
| Does consent prevent CELLS DERIVED FROM THE DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
Does consent permit research by | |
| an academic institution? | Yes |
| a public organisation? | Yes |
| a non-profit company? | Yes |
| a for-profit corporation? | No |
| How may genetic information associated with the cell line be accessed? | Open Access |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Does the consent anticipate that the donor will be notified of results or outcomes of any research involving the donated samples or derived cells? | No |
| Does the consent permit the donor, upon withdrawal of consent, to stop the use of the derived cell line(s) that have already been created from donated samples? | No |
| Does the consent permit the donor, upon withdrawal of consent, to stop delivery or use of information and data about the donor? | No |
| Does consent permit access to medical records of the donor? | No |
| Does consent permit access to any other source of information about the clinical treatment or health of the donor? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | HTA, HFEA, and UK Stem Cell Bank, Central Manchester LREC favourable opinion 03/CM/684 |
| Approval number | HTA Licence number: 22627 HFEA Licence number: R0171-4-a. UKSCB approval number: SCSC12-28 |
hESC Derivation
| Date of derivation | 2011-09-05 |
| Embryo stage | Blastula with ICM and Trophoblast |
| Supernumerary embryos from IVF treatment? |
Yes
Separation of research and IVF treatment?
Yes |
| PGD Embryo? |
No |
| Expansion status |
2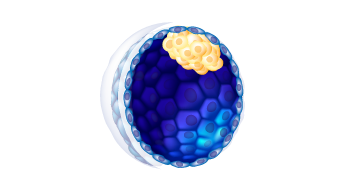
|
| ICM morphology |
D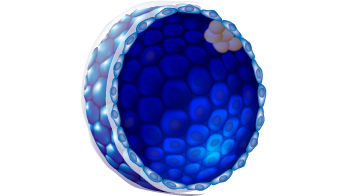
|
| Trophectoderm morphology |
a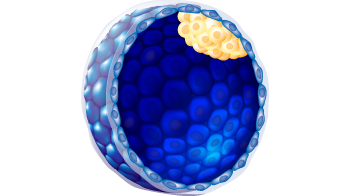
|
| ZP removal technique | Chemical |
| Cell isolation | Mechanical |
| Cell seeding | Isolated ICM |
| Derived under xeno-free conditions? |
No |
| Derivation under GMP? |
Yes |
| Available as clinical grade? |
Yes |
Culture Conditions
| Surface coating | Cellstart |
| Feeder cells |
Human dermal fibroblast Cellfinder Ont Id: CELDA_000001419 |
| Passage method | Mechanically |
| O2 Concentration | 20 % |
| CO2 Concentration | 5 % |
| Medium |
mTeSR™ 2
|
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| NANOG |
Yes |
|
||||
| POU5F1 (OCT-4) |
Yes |
|
|
|||
| SOX2 |
Yes |
|
||||
| SSEA-1 |
No |
|
||||
| SSEA-3 |
Yes |
|
||||
| SSEA-4 |
Yes |
|
||||
| TRA 1-60 |
Yes |
|
||||
| TRA 1-81 |
Yes |
|
Data on pluripotency is in Ye et al 2017 DOI 10.1186/s13287-017-0561-y
Differentiation Potency
In vitro spontaneous differentiation
| Marker | Expressed |
| GATA Binding Protein 6 |
Yes |
| FOXA2 |
Yes |
In vitro spontaneous differentiation
| Marker | Expressed |
| Vimentin |
Yes |
| Actin, Alpha 2, Smooth Muscle, Aorta |
Yes |
In vitro spontaneous differentiation
| Marker | Expressed |
| Tubulin Beta 3 Class III |
Yes |
| Neurofilament, light polypeptide |
Yes |
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
| Mycoplasma | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
Trnscriptome
Transcriptome as part of a disease modeling study |


Login to share your feedback, experiences or results with the research community.