SOD1-G128R_K7-4L-f, SOD1-G128R
ICGi022-A-1
General
Cell Line |
|
| hPSCreg name | ICGi022-A-1 |
| Cite as: | ICGi022-A-1 (RRID:CVCL_D0VR) |
| Alternative name(s) |
SOD1-G128R_K7-4L-f, SOD1-G128R
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
WAe009-A-1P (IsoHD-30Q-NGN2) WAe009-A-1Q (IsoHD-65Q-NGN2) GIBHi002-A-1 (C5-PARK2-KO) WAe009-A-1R (IsoHD-81Q-NGN2) STBCi026-A-1 (SFC840-03-03 LRRK2-/-D10) STBCi026-A-2 (SFC840-03-03 LRRK2-/-C11) STBCi026-A-3 (SFC840-03-03 LRRK2 WT/R1441C H3) SIGi001-A-13 (iPSC0028 – MonoAllelic MAPT_Ex10+16T/Clone 1D01-11) BIONi037-A-2 (BIONi037-A ApoE2/2 #M10-7) BIONi037-A-3 (BIONi037-A ApoE3/4 #P10-22) BIONi037-A-4 (BIONi037-A ApoE4/4 #I10-53) BIONi037-A-1 (16423 ApoE KO) |
| Last update | 24th June 2022 |
| User feedback | |
Provider |
|
| Generator | Institute of Cytology and Genetics, Siberian Branch of Russian Academy of Sciences (ICG) |
External Databases |
|
| BioSamples | SAMEA9333558 |
| Cellosaurus | CVCL_D0VR |
| Wikidata | Q123032655 |
General Information |
|
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: not allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | female |
| Ethnicity | Caucasian |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Disease associated phenotypes | no phenotypes |
Karyotyping (Donor) |
|
| Has the donor karyotype been analysed? |
Unknown
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
Exome sequencing
https://www.ncbi.nlm.nih.gov/sra/?term=SRR11413027 No disease associated mutation found |
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMN14446266 |
Ethics
Also have a look at the ethics information for the parental line
ICGi022-A
.
| Is there an MTA available for the cell line? | No |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? | IDT crRNA and tracrRNA were used for CRISPR/Cas9 gene editing |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hIPSC Derivation
General |
|
|
The source cell information can be found in the parental cell line
ICGi022-A.
|
|
| Passage number reprogrammed | 1 |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Episomal |
| Genes | |
| Is reprogramming vector detectable? |
No |
| Methods used |
PCR
|
| Files and images showing reprogramming vector expressed or silenced | |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Selection criteria for clones | ESC-like morphology |
| Derived under xeno-free conditions |
No |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Gelatin | |||||||||||||||
| Feeder cells |
Mouse embryonic fibroblasts Cellfinder Ont Id: http://www.ebi.ac.uk/efo/EFO_0004040 |
|||||||||||||||
| Passage method |
Enzymatically
TrypLE
|
|||||||||||||||
| O2 Concentration | 20 % | |||||||||||||||
| CO2 Concentration | 5 % | |||||||||||||||
| Medium |
Other medium:
Base medium: Knock-out DMEM
Main protein source: Knock-out serum replacement Serum concentration: 15 % Supplements
|
|||||||||||||||
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
|||||||||||||||
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
|||||||||||||||
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
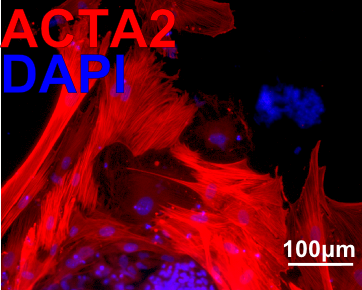
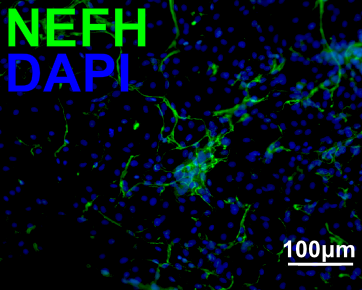
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| NANOG |
Yes |
|||||
| SOX2 |
Yes |
|||||
| POU5F1 (OCT-4) |
Yes |
|||||
| TRA 1-60 |
Yes |
Score:
| Marker | Present | Absent |
| mCpG | ||
| OCT4 |
Method documentation
Differentiation Potency
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Disease/phenotype related modifications |
|
| Genetic modifications not related to a disease |
|




Login to share your feedback, experiences or results with the research community.