CN090 C1B5B5
UPITTi004-B
General
Cell Line |
|
| hPSCreg name | UPITTi004-B |
| Cite as: | UPITTi004-B (RRID:CVCL_D0P7) |
| Alternative name(s) |
CN090 C1B5B5
|
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
RNRMUi005-A (EB-iPSC-d4, RDEB-iPSC-d4) Donor diseases: Epidermolysis Bullosa Simplex recessive dystrophic epidermolysis bullosa UNIZARi001-A (FiPSTK2-2) Donor's gene variants: TK2, TK2 Donor diseases: Mitochondrial DNA depletion syndrome, myopathic form HDZi003-A (hiPSC NP0038) Donor's gene variants: TMEM43 Donor diseases: arrhythmogenic right ventricular dysplasia 5 HIHDNDi001-A (A30P-3, SNCA3, Tue_020_A) Donor's gene variants: SNCA, SNCA, SNCA Donor diseases: autosomal dominant Parkinson disease 1 HIHDNDi001-B (A30P-4, SNCA4, Tue_020_B) Donor's gene variants: SNCA, SNCA, SNCA Donor diseases: autosomal dominant Parkinson disease 1 UNIFEi002-A (UNIFEi013-A, UniFei013-A, MARPiPSCA1N13, iPSca1N13) Donor diseases: Spinocerebellar Ataxia Type 1 |
| Last update | 16th August 2023 |
| User feedback | |
Provider |
|
| Generator | University of Pittsburgh (UPITT) |
External Databases |
|
| BioSamples | SAMEA114066531 |
| Cellosaurus | CVCL_D0P7 |
| Wikidata | Q123033756 |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: allowed
|
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Black |
Phenotype and Disease related information (Donor) |
|
| Diseases | A disease was diagnosed.
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA114066391 |
Ethics
| Has informed consent been obtained from the donor of the embryo/tissue from which the pluripotent stem cells have been derived? | Yes |
| Was the consent voluntarily given? | Yes |
| Has the donor been informed that participation will not directly influence their personal treatment? | Yes |
| Can you provide us with a copy of the Donor Information Sheet provided to the donor? | Yes |
| Do you (Depositor/Provider) hold the original Donor Consent Form? | No |
| If you do not hold the Donor Consent Form, do you know who does? | Yes |
| Please indicate whether the data associated with the donated material has been pseudonymised or anonymised. | pseudonymised |
| Does consent explicitly allow the derivation of pluripotent stem cells? | No |
| Does consent prevent CELLS DERIVED FROM THE DONATED BIOSAMPLE from being made available to researchers anywhere in the world? | No |
| How may genetic information associated with the cell line be accessed? | Controlled Access |
| Will the donor expect to receive financial benefit, beyond reasonable expenses, in return for donating the biosample? | No |
| Has a favourable opinion been obtained from a research ethics committee, or other ethics review panel, in relation to the Research Protocol including the consent provisions? | Yes |
| Name of accrediting authority involved? | University of Pittsburgh IRB & clinicaltrials.gov |
| Approval number | STUDY19110083 & NCT02972801 |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? |
hIPSC Derivation
General |
|
| Source cell type |
Synonyms
|
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Sendai virus |
| Is reprogramming vector detectable? |
No |
| Methods used |
RT-PCR
|
Vector free reprogramming |
|
Other |
|
| Derived under xeno-free conditions |
Unknown |
| Derived under GMP? |
Unknown |
| Available as clinical grade? |
Unknown |
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Medium |
mTeSR™ Plus
|
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| DPPA2 |
Yes |
|||||
| NANOG |
Yes |
|
||||
| SSEA-4 |
Yes |
|
||||
| TRA 1-81 |
Yes |
|
||||
| SOX2 |
Yes |
|
|
|||
| POU5F1 (OCT-4) |
Yes |
|
|
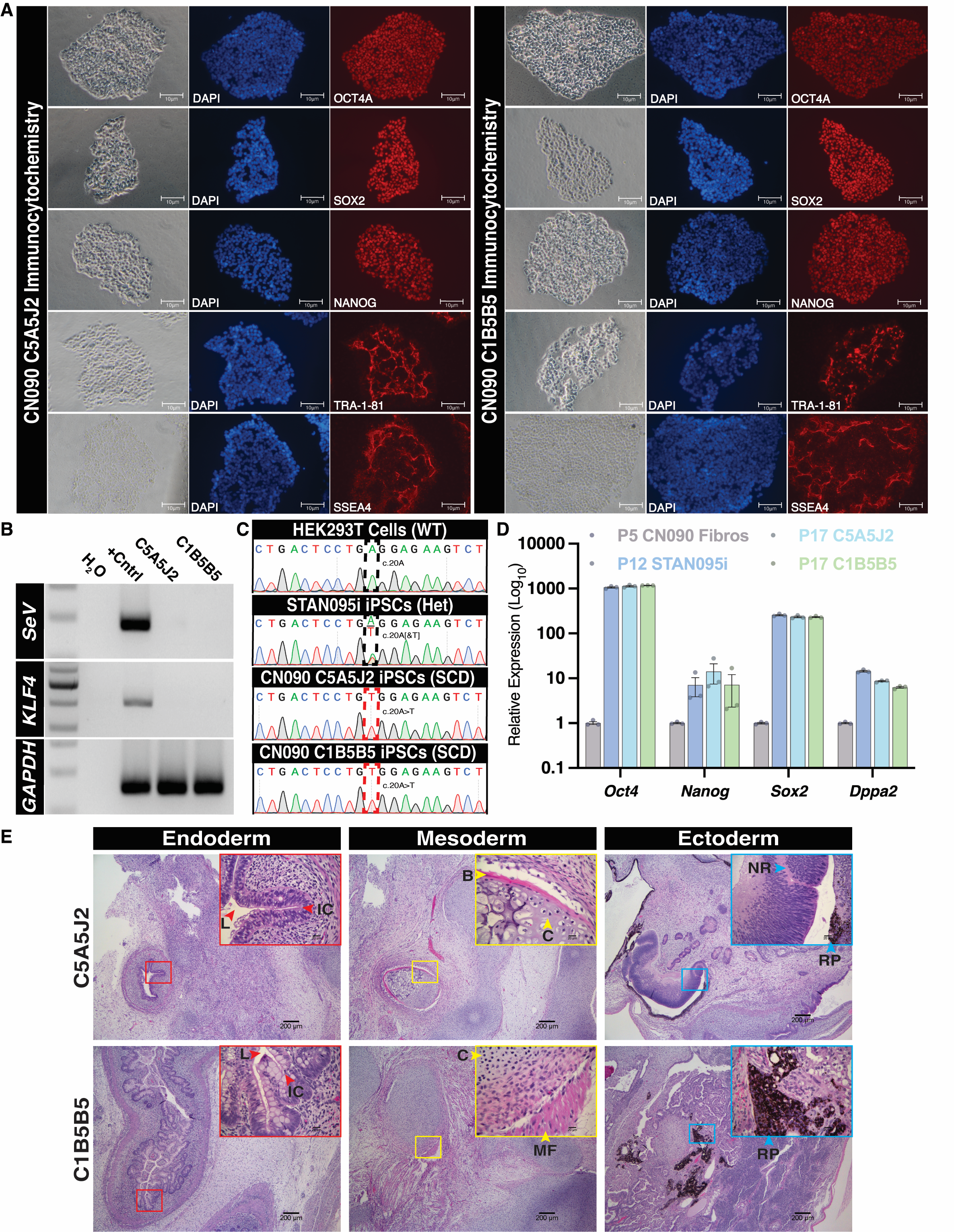
Teratoma analysis shows a robust, non-cancerous, tri-lineage differential potential. Teratomas were processed and analysed with a histopathologist and show definitive differentiation for each germ layer (Fig. 1E). Endodermal differentiation in both lines presented a strong gastrointestinal phenotype, indicated by the presence of a lumen (L) and intestinal crypts (IC) (left). Mesoderm differentiation is indicated by cartilage (C) and bone (B) in C5A5J2 (middle, top), and by cartilage and muscle fibers (MF) in C1B5B5 (middle, bottom). Ectoderm differentiation in both lines presented with retinal pigment (RP), and in C5A5J2, a neural rosette (NR) was also observed (top, right).
Method documentation
Differentiation Potency
In vivo teratoma
Morphology
C1B5B5 Teratoma Mouse 1.pdf
Endoderm is bottom row.
C1B5B5 Teratoma Mouse 2.pdf
Endoderm is bottom row.
C1B5B5 Teratoma Mouse 3.pdf
Endoderm is bottom row.
Protocol or reference
In vivo teratoma
Morphology
C1B5B5 Teratoma Mouse 1.pdf
Mesoderm is middle row.
C1B5B5 Teratoma Mouse 2.pdf
Mesoderm is middle row.
C1B5B5 Teratoma Mouse 3.pdf
Mesoderm is middle row.
Protocol or reference
In vivo teratoma
Morphology
C1B5B5 Teratoma Mouse 1.pdf
Ectoderm is top row.
C1B5B5 Teratoma Mouse 2.pdf
Ectoderm is top row.
C1B5B5 Teratoma Mouse 3.pdf
Ectoderm is top row.
Protocol or reference
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
46XY
|
Other Genotyping (Cell Line) |
|


Login to share your feedback, experiences or results with the research community.