WAe001-A-2F
General
Cell Line |
|
| hPSCreg name | WAe001-A-2F |
| Cite as: | WAe001-A-2F (RRID:CVCL_D6SS) |
| Cell line type | Human embryonic stem cell (hESC) |
| Similar lines |
WAe001-A (WA01, H1) WAe001-A-A (H1X-10/H1.2-20) WAe001-A-2B (H1-NSD2-/-) WAe001-A-B (H1X-59/H1.2-6) WAe001-A-C (H1X-10/H1.3-62) WAe001-A-D (H1X-59/H1.3-68) WAe001-A-E (H1X-10/H1.4-4) WAe001-A-F (H1X-59/H1.4-5) WAe001-A-86 (H1.139, H1.1-39) WAe001-A-72 (H1-FTO-Knockout-13) WAe001-A-31 (H1_RB1ex1_D6) WAe001-A-32 (H1_RB1ex1_E9) WAe001-A-87 (H1.213, H1.2-13) WAe001-A-G (H1X-10/H1.5-1) WAe001-A-88 (H1.2-124, H1.2124) WAe001-A-1N (H1-FIS1-KO, WA01-FIS1-KO) WAe001-A-89 (H1.3-64, H1.364) WAe001-A-H (H1X-59/H1.5-33) WAe001-A-1O (WA01-MYB-KO, H1-MYB-KO) WAe001-A-77 (H1 RYBP-FLAG-HA KI) WAe001-A-90 (H1.3-191, H1.3191) WAe001-A-I (H1.0-29/H1.1-13) WAe001-A-41 (H1-RNF2-KO-1#) WAe001-A-J (H1.0-102/H1.1-34) WAe001-A-42 (H1-RNF2-KO-2#) WAe001-A-K (H1.0-29/H1.2-31) WAe001-A-1 (H1.1, WA01.1) WAe001-A-L (H1.0-102/H1.2-6) WAe001-A-M (H1.0-29/H1.3-22) WAe001-A-N (H1.0-102/H1.3-90) WAe001-A-91 (H1.4-126, H1.4126) WAe001-A-O (H1.0-29/H1.4-14) WAe001-A-92 (H1.4-143, H1.4143) WAe001-A-P (H1.0-102/H1.4-19) WAe001-A-93 (H1.5-23, H1.523) WAe001-A-78 (H1X10, H1X-10) WAe001-A-94 (H1.528, H1.5-28) WAe001-A-Q (H1.0-29/H1.5-6) WAe001-A-73 (H1-GATAD2B-KO-79#) WAe001-A-R (H1.0-102/H1.5-44) WAe001-A-10 (H1_RB1ex3_G3) WAe001-A-74 (H1-GATAD2B-KO-81#) WAe001-A-S (H1.1-17/H1.2-11) WAe001-A-11 (H1_RB1ex3_G4) WAe001-A-T (H1.1-39/H1.2-9) WAe001-A-U (H1.1-17/H1.4-18) |
| Last update | 22nd February 2024 |
| User feedback | |
Provider |
|
| Generator |
Hainan Medical University (HNMU)
Contact:
Hainan Medical University (HNMU) |
| Owner | Hainan Medical University (HNMU) |
| Distributors | |
| Derivation country | China |
External Databases |
|
| BioSamples | SAMEA115343060 |
| Cellosaurus | CVCL_D6SS |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | N/A |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Family history | NO |
| Is the medical history available upon request? | NO |
| Is clinical information available? | NO |
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
| All cell lines of this donor's relatives |
Has brother:
|
External Databases (Donor) |
|
| BioSamples | SAMEA7767418 |
Ethics
Also have a look at the ethics information for the parental line
WAe001-A
.
| Is there an MTA available for the cell line? | No |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hESC Derivation
|
The source cell information can be found in the parental cell line
WAe001-A.
|
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
EDTA
|
| O2 Concentration | 21 % |
| CO2 Concentration | 5 % |
| Medium |
mTeSR™ 1
|
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | No |
Characterisation
Analysis of Undifferentiated Cells
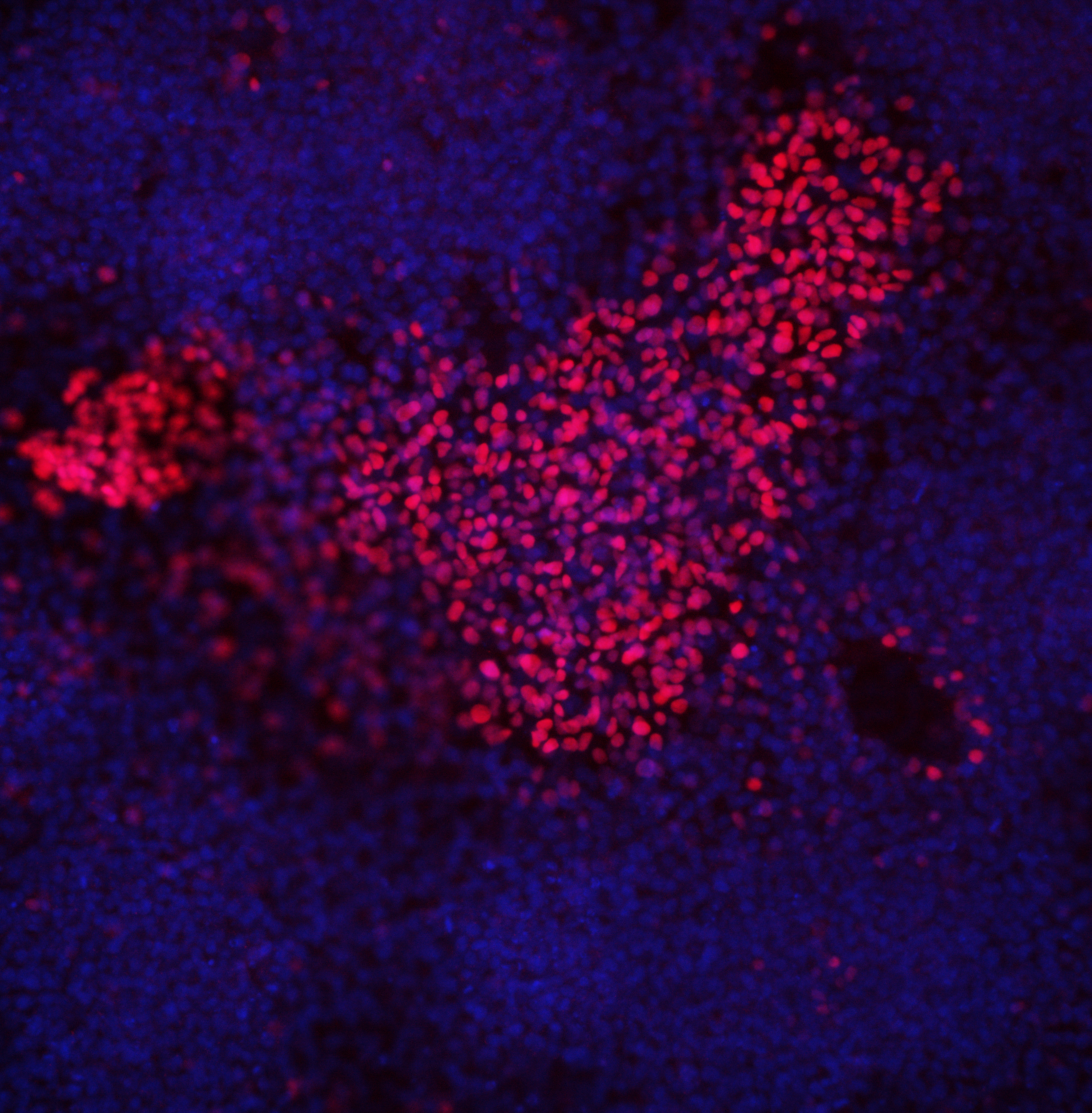
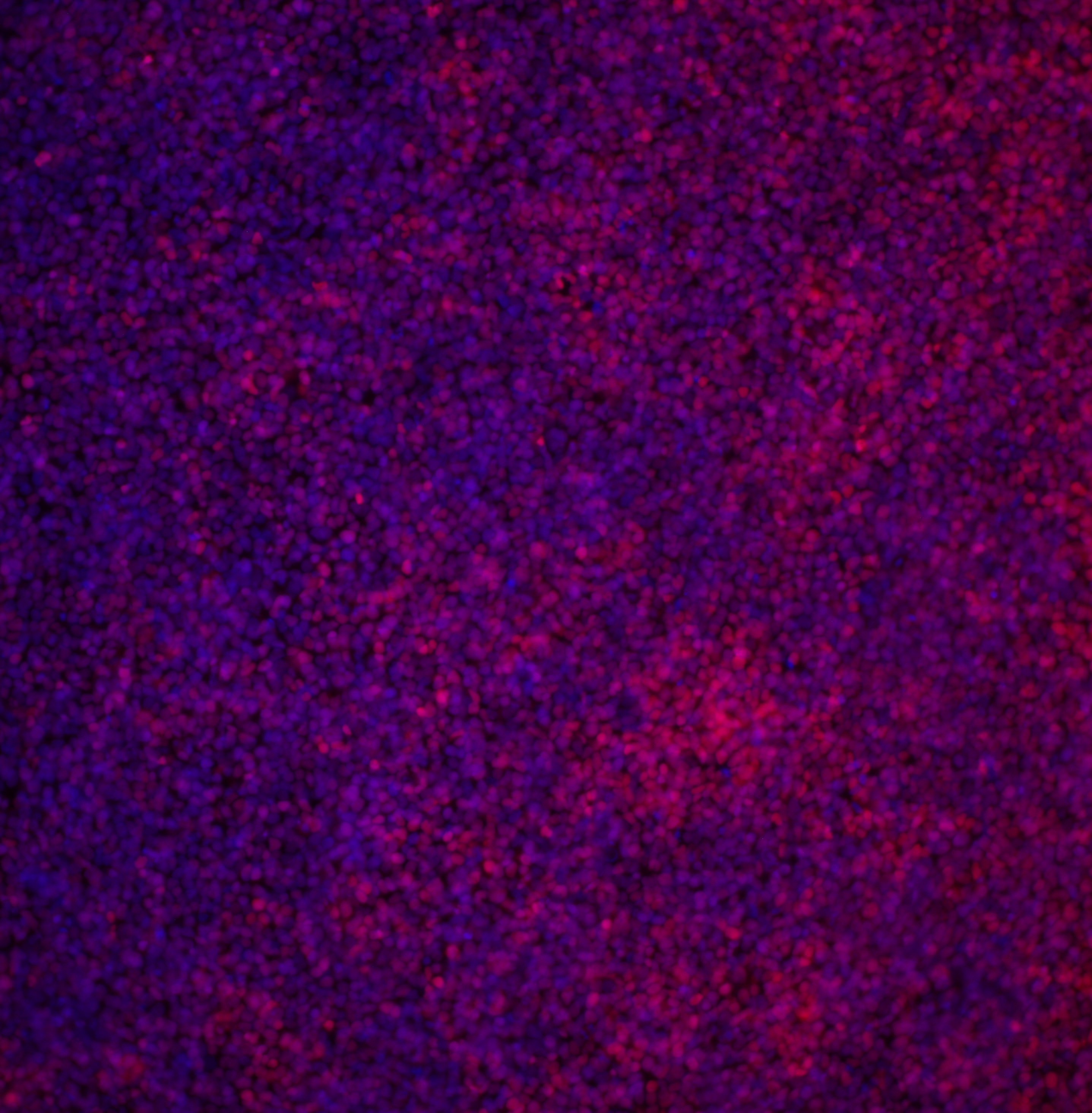
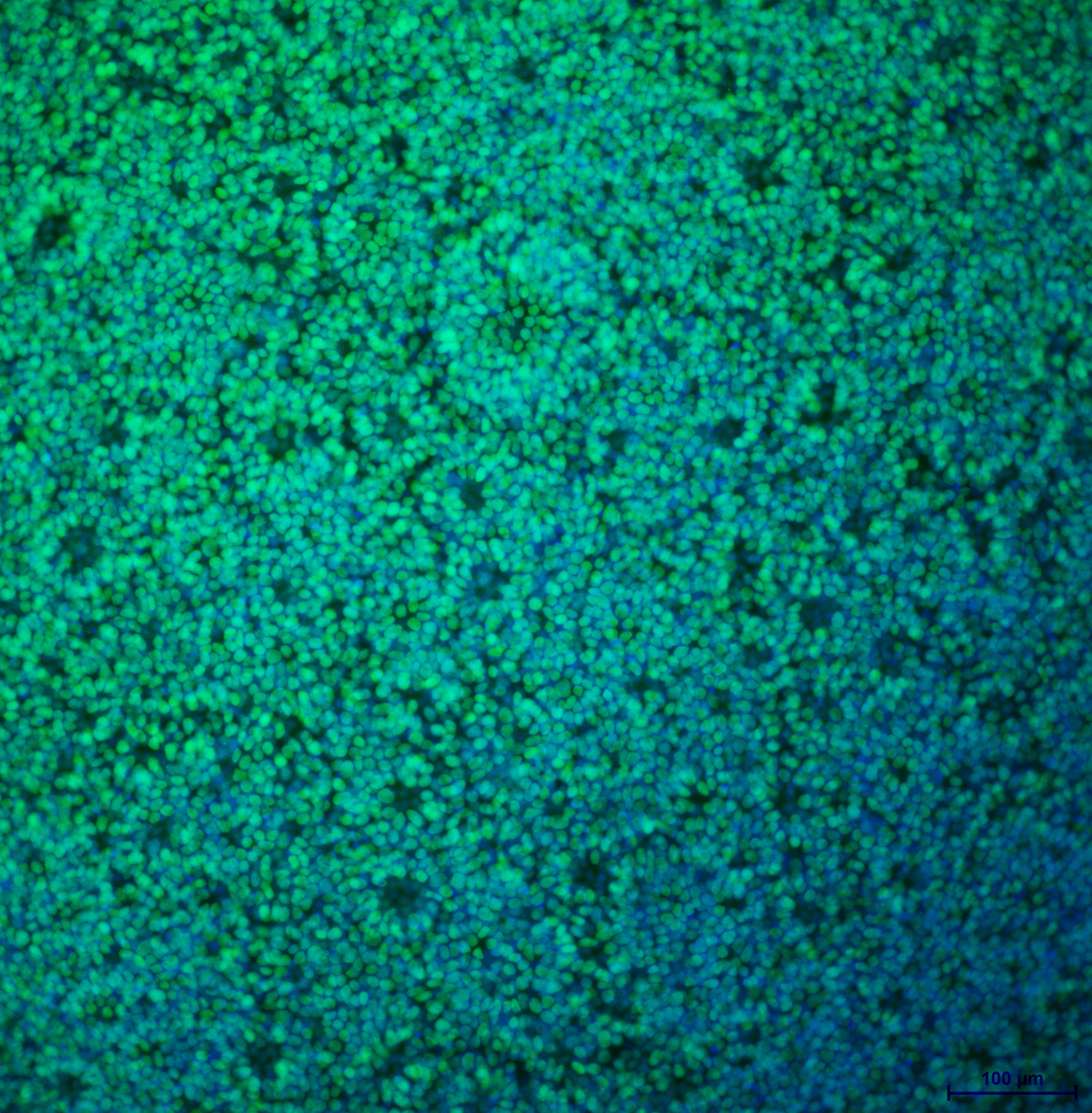
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| SOX2 |
Yes |
|||||
| POU5F1 (OCT-4) |
Yes |
|||||
| SSEA-4 |
Yes |
|||||
| TRA 1-60 |
Yes |
Score:
| Marker | Present | Absent |
| mCpG | ||
| OCT4 |
Differentiation Potency
Microbiology / Virus Screening |
|
| Mycoplasma | Negative |
Certificate of Analysis |
|
| Is there a certificate of analysis available? |
Yes
Passage:
|
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Disease/phenotype related modifications |
|
| Genetic modifications not related to a disease |
|


Login to share your feedback, experiences or results with the research community.