SCTi003-A-2
General
Cell Line |
|
| hPSCreg name | SCTi003-A-2 |
| Cite as: | SCTi003-A-2 |
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
SIGi001-A-13 (iPSC0028 – MonoAllelic MAPT_Ex10+16T/Clone 1D01-11) BIONi037-A-2 (BIONi037-A ApoE2/2 #M10-7) BIONi037-A-3 (BIONi037-A ApoE3/4 #P10-22) BIONi037-A-4 (BIONi037-A ApoE4/4 #I10-53) GEMi011-A-1 (GN-JY02-02-011(PSEN1-exon9-KO)) HMGUi001-A-44 (BIHi043-A-SORL_SNP_Cl-2) BIONi037-A-1 (16423 ApoE KO) WIBRe001-A-58 (WIBR3_VPS13C_FS_Homo_H3-1) WIBRe001-A-59 (WIBR3_VPS13C_A444P_Het_E12-1) WIBRe001-A-60 (WIBR3_VPS13C_A444P_Homo_C8-2) WIBRe001-A-5 (WIBR3_PRKN_X3DEL_F2-5) WIBRe001-A-6 (WIBR3_PRKN_X3DEL_H2-2) WIBRe001-A-7 (WIBR3_SNCA_A30P_A2-3) WIBRe001-A-8 (WIBR3_SNCA_A30P_E1-3) WIBRe001-A-9 (WIBR3_FBXO7_R498X_A7-1) WIBRe001-A-10 (WIBR3_FBXO7_FS_A3-1) WIBRe001-A-11 (WIBR3_PINK1_Q129X_C4-1) WIBRe001-A-12 (WIBR3_PINK1_Q129X_E2-2) WIBRe001-A-13 (WIBR3_PINK1_Q129X_E7-1) WIBRe001-A-15 (WIBR3_DNAJC6_c.801-2 A>G+FS/c.801-2 A>G_G12-2) WIBRe001-A-16 (WIBR3_DNAJC6_FS/FS_H10-1) WIBRe001-A-17 (WIBR3_GBA1_IVS2_G2E) WIBRe001-A-18 (WIBR3_GBA1_IVS2_E10B) WIBRe001-A-19 (WIBR3_SNCA_A53T_1) WIBRe001-A-20 (WIBR3_SNCA_A53T_2) WIBRe001-A-21 (WIBR3_SNCA_A53T_4) WIBRe001-A-22 (WIBR3_LRRK2_G2019S_5_Het) WIBRe001-A-23 (WIBR3_LRRK2_G2019S_6_Het) WIBRe001-A-24 (WIBR3_LRRK2_G2019S_216_Het) WIBRe001-A-25 (WIBR3_LRRK2_G2019S_65_Homo) WIBRe001-A-26 (WIBR3_DJ1_X1-5DEL_2860) WIBRe001-A-27 (WIBR3_DJ1_X1-5DEL_2872) WIBRe001-A-28 (WIBR3_DJ1_X1-5DEL_2876) WIBRe001-A-74 (WIBR3_FBXO7_R498X_A7-1) WIBRe001-A-61 (WIBR3_DJ1_X1-5DEL_Het_2036) WIBRe001-A-62 (WIBR3_DJ1_X1-5DEL_Het_2038) WIBRe001-A-32 (WIBR3_EWT_S1) |
| Last update | 26th August 2025 |
| Notes | Genome-edited Human hiPSC Line, SCTi003-A-2, APP K670N/M671L (Swedish Mutation) (SCTi003-A-2) was generated by CRISPR/Cas9 technology, which introduced the amyloid precursor protein (APP) K670N/M671L double mutation (also known as the Swedish mutation, c.2026A>G & c.2032T>A) into the endogenous APP locus. The Swedish mutation is a well-characterized familial Alzheimer’s disease mutation associated with increased β-secretase cleavage of APP, leading to an elevated production of amyloid-β peptides and early-onset Alzheimer’s pathology (Sasaguri et al.; Thordardottir & Graff).
This product’s parental human induced pluripotent stem cell (hiPSC) line, Healthy Control Human iPSC Line, Female, SCTi003-A, is a well-characterized control line derived from peripheral blood mononuclear cells (PBMCs) from a 48-year-old donor. Targeted gene modifications were confirmed by Sanger sequencing. Post-editing, extensive quality control procedures were undertaken in the manufacturing process for SCTi003-A-2 to ensure optimal product performance and reproducibility. SCTi003-A-2 is karyotypically stable, expresses markers of the undifferentiated state, and remains capable of directed differentiation into all three germ layers, including neural lineage cells relevant for Alzheimer’s disease research. This genome-edited hiPSC line, along with its parental hiPSC line, provides a robust, isogenic disease modeling tool for studying amyloidogenic processing, studying Alzheimer’s disease mechanisms, and evaluating therapeutic interventions targeting the APP pathway. SCTi003-A-2 is manufactured with mTeSR™ Plus (Catalog #100-0276) and is compatible with STEMdiff™ cell culture media products, allowing for standardized high-quality maintenance and differentiation to various cell types, such as neurons, astrocytes, and microglia. Cells were obtained using Institutional Review Board (IRB)-approved consent forms and protocols. |
| User feedback | |
Provider |
|
| Generator |
STEMCELL Technologies Inc. (SCT)
Contact:
STEMCELL Technologies Inc. (SCT) |
| Owner | STEMCELL Technologies Inc. (SCT) |
| Distributors | |
| Derivation country | United States |
External Databases |
|
| BioSamples | SAMEA119562656 |
General Information |
|
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | female |
| Ethnicity |
Self-declared race/ethnicity = White Ancestry = 100% European |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Is the medical history available upon request? | No |
| Is clinical information available? | No |
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
External Databases (Donor) |
|
| BioSamples | SAMEA11371632 |
Ethics
Also have a look at the ethics information for the parental line
SCTi003-A
.
| Is there an MTA available for the cell line? | No |
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? | |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hIPSC Derivation
General |
|
|
The source cell information can be found in the parental cell line
SCTi003-A.
|
|
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Proprietary non-integrating reprogramming technology |
| Is reprogramming vector detectable? |
No |
| Methods used |
PCR
|
| Notes on reprogramming vector detection | Clearance confirmed at passage 21 |
Vector free reprogramming |
|
| Type of used vector free reprogramming factor(s) |
None
|
Other |
|
| Derived under xeno-free conditions |
No |
| Derived under GMP? |
No |
| Available as clinical grade? |
No |
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
ReLeSR™
|
| O2 Concentration | 20 % |
| CO2 Concentration | 5 % |
| Medium | mTeSR™ Plus |
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | No |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | No |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| TRA 1-60 |
Yes |
Score:
| Marker | Present | Absent |
| mCpG | ||
| OCT4 |
Morphology pictures
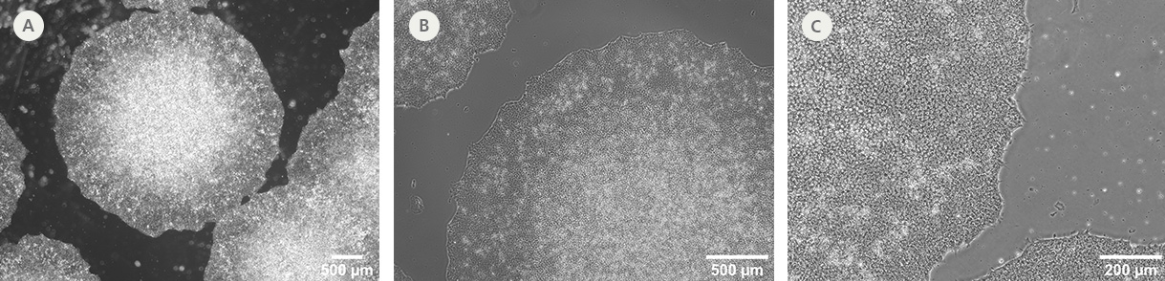
Figure 2. SCTi003-A-2 Human iPSCs Demonstrate High-Quality Morphology in Routine Culture.
Cryopreserved cells from line SCTi003-A-2 were thawed and maintained in mTeSR™ Plus (Catalog #100-1130) on Corning® Matrigel® Matrix. (A) The resulting iPSC colonies have densely packed cells and show multi-layering when ready to be passaged. (B,C) Cells retain prominent nucleoli and high nuclear-to-cytoplasmic ratios. iPSC = induced pluripotent stem cell.
Cryopreserved cells from line SCTi003-A-2 were thawed and maintained in mTeSR™ Plus (Catalog #100-1130) on Corning® Matrigel® Matrix. (A) The resulting iPSC colonies have densely packed cells and show multi-layering when ready to be passaged. (B,C) Cells retain prominent nucleoli and high nuclear-to-cytoplasmic ratios. iPSC = induced pluripotent stem cell.
Differentiation Potency
In vitro directed differentiation
Protocol or reference
In vitro directed differentiation
Protocol or reference
In vitro directed differentiation
Protocol or reference
Microbiology / Virus Screening |
|
| HIV 1 | Negative |
| HIV 2 | Negative |
| Hepatitis B | Negative |
| Hepatitis C | Negative |
| Mycoplasma | Negative |
Certificate of Analysis |
|
| Is there a certificate of analysis available? |
Yes
Passage:
37
|
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
| Is there genome-wide genotyping or functional data available? |
Yes
Exome sequencing
Whole exome sequencing data file (Catalog #500-0707) is available upon request for a fee for SCTi003-A-2 customers. Please contact iPSCrequests@stemcell.com for more information.
SNP typing array
SNP microarray data is included in each lot-specific COA. Please contact iPSCrequests@stemcell.com for more information.
Genome Sequencing
Whole genome sequencing data file (Catalog #500-0708) is available upon request for a fee for SCTi003-A-2 customers. Please contact iPSCrequests@stemcell.com for more information. |
Genetic Modification
| Disease/phenotype related modifications |
|


Login to share your feedback, experiences or results with the research community.