BIONi010-C-72
General
Cell Line |
|
| hPSCreg name | BIONi010-C-72 |
| Cite as: | BIONi010-C-72 (RRID:CVCL_D6RU) |
| Cell line type | Human induced pluripotent stem cell (hiPSC) |
| Similar lines |
BIONi010-C (BIONi010-C, K3P53) BIONi010-C-25 (BIONi010-C heterozygous TREM2 KO) BIONi010-C-2 (BIONi010-C ApoE E3/E3 #H8 P32) BIONi010-C-3 (BIONi010-C ApoE KO #KO30 P30) BIONi010-C-70 (BIONi010-C with an APOE 2/2 genotype with an additional, homozygous christchurch mutation) BIONi010-C-71 (BIONi010-C with an APOE 3/3 genotype with an additional, homozygous christchurch mutation) BIONi010-C-4 (BIONi010-C ApoE E4/E4 #B44 P27) BIONi010-C-5 (BIONi010-C CD33 E2del #N14 P26) BIONi010-C-6 (BIONi010-C ApoE E2/E2) BIONi010-C-7 (BIONi010-C Trem2 R47H) BIONi010-C-8 (BIONi010-C Trem2 T66M, #Y5-80) BIONi010-C-9 (BIONi010-C CD33 KO) BIONi010-C-17 (BIONi010-C TREM2 KO) BIONi010-C-56 (BIONi010-C-A713T-C25) BIONi010-C-57 (BIONi010-C-A713T-C42) BIONi010-C-58 (BIONi010-C-A713T-C1) BIONi010-C-59 (BIONi010-C-A713T-C33) BIONi010-C-60 (BIONi010-C-R589C-C7) BIONi010-C-61 (BIONi010-C-R589C-C16) BIONi010-C-62 (BIONi010-C-R589C-C5) BIONi010-C-63 (BIONi010-C-R589C-C9) BIONi010-C-18 (BIONi010-C TBK1 KO) BIONi010-C-19 (BIONi010-C IKBKE KO) BIONi010-C-43 (BIONi010-C + aSNCA-wt AAVS1) BIONi010-C-44 (BIONi010-C + aSNCA-A53T AAVS1) BIONi010-C-10 (HNF1AP291fsinsC +/- 54-5) BIONi010-C-11 (HNF1AP291fsinsC -/- 66-1) BIONi010-C-12 (HNF4ApR309C -/- 2-4) BIONi010-C-52 (BIONi010-C with an APOE 2/2 genotype (with two functional alleles in contrast to BIONi010-C-6)) BIONi010-C-53 (BIONi010-C with an APOE 3/3 genotype (with two functional alleles in contrast to BIONi010-C-2)) BIONi010-C-54 (BIONi010-C with an APOE 4/4 genotype (with two functional alleles in contrast to BIONi010-C-4)) BIONi010-C-48 (BIONi010-C hMDR1) BIONi010-C-45 (BIONi010-C iCRE AAVS1 GBA1 LoxP EX5-6) BIONi010-C-64 (BIONi010-C-T2A-Nanoluciferase reporter cl. 29) BIONi010-C-51 (BIONi010-C TNNI3-mCherry reporter) BIONi010-C-13 (BIONi010-C + NGN2 #I7-26) BIONi010-C-15 (BIONi010-C +dox inducible NGN2-GFP) BIONi010-C-55 (BIONi010-C TNNI3-mCherry/TNNI1-EGFP dual reporter cl. 74) BIONi010-C-41 (BIONi010-C + iNGN2 Two-plasmid system/CRISPR-2) BIONi010-C-42 (BIONi010-C + iCRE AAVS1) BIONi010-C-49 (BIONi010-C + synapsin-m2rtTA + SNCA-wt) BIONi010-C-50 (BIONi010-C + synapsin-m2rtTA + SNCA-A53T) BIONi010-C-24 (BIONi010-C Dox a-syn) BIONi010-C-65 (BiONI010-C-O16) BIONi010-C-66 (BIONi010-C-N7) BIONi010-A (K1P53) BIONi010-B (K2P53, BIONi010-B) |
| Last update | 30th January 2024 |
| Notes | The line has an APOE 4/4 genotype with an additional, homozygous christchurch mutation |
| User feedback | |
Provider |
|
| Generator |
Bioneer (BION)
Contact:
Bioneer (BION) |
| Owner | Bioneer (BION) |
| Distributors | |
| Derivation country | Denmark |
External Databases |
|
| BioSamples | SAMEA115162597 |
| Cellosaurus | CVCL_D6RU |
General Information |
|
| Publications | |
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: not allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | male |
| Ethnicity | Black or African-American |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA3105780 |
Ethics
Also have a look at the ethics information for the parental line
BIONi010-C
.
| For generation of the cell line, who was the supplier of any recombined DNA vectors or commercial kits used? |
hIPSC Derivation
General |
|
|
The source cell information can be found in the parental cell line
BIONi010-C.
|
|
| Passage number reprogrammed | 32 |
Reprogramming method |
|
| Vector type | Non-integrating |
| Vector | Episomal |
| Methods used |
PCR
|
Vector free reprogramming |
|
Other |
|
| Derived under xeno-free conditions |
Unknown |
| Derived under GMP? |
Unknown |
| Available as clinical grade? |
Unknown |
Culture Conditions
| Medium |
mTeSR™ Plus
|
Characterisation
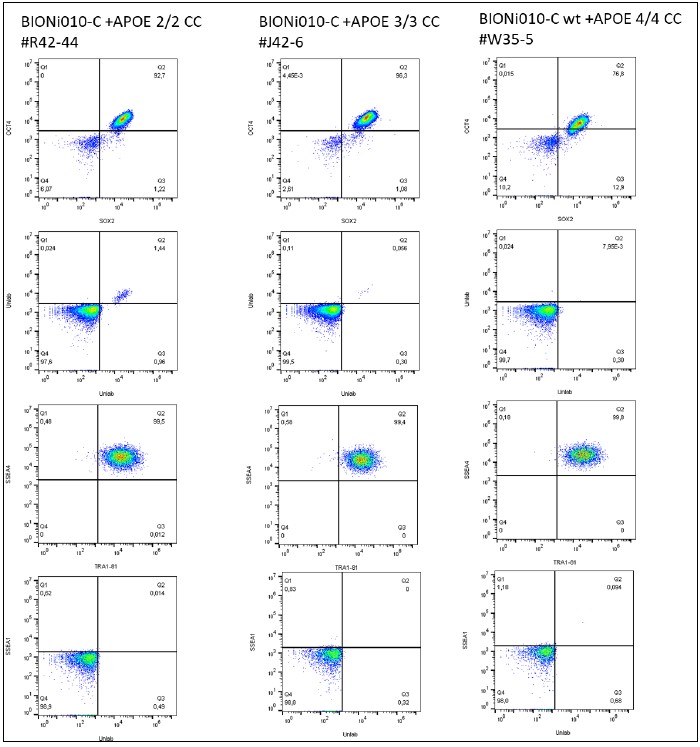
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| SOX2 |
Yes |
|
||||
| TRA 1-81 |
Yes |
|
||||
| SSEA-1 |
No |
|
||||
| SSEA-4 |
Yes |
|
Differentiation Potency
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Disease/phenotype related modifications |
|


Login to share your feedback, experiences or results with the research community.