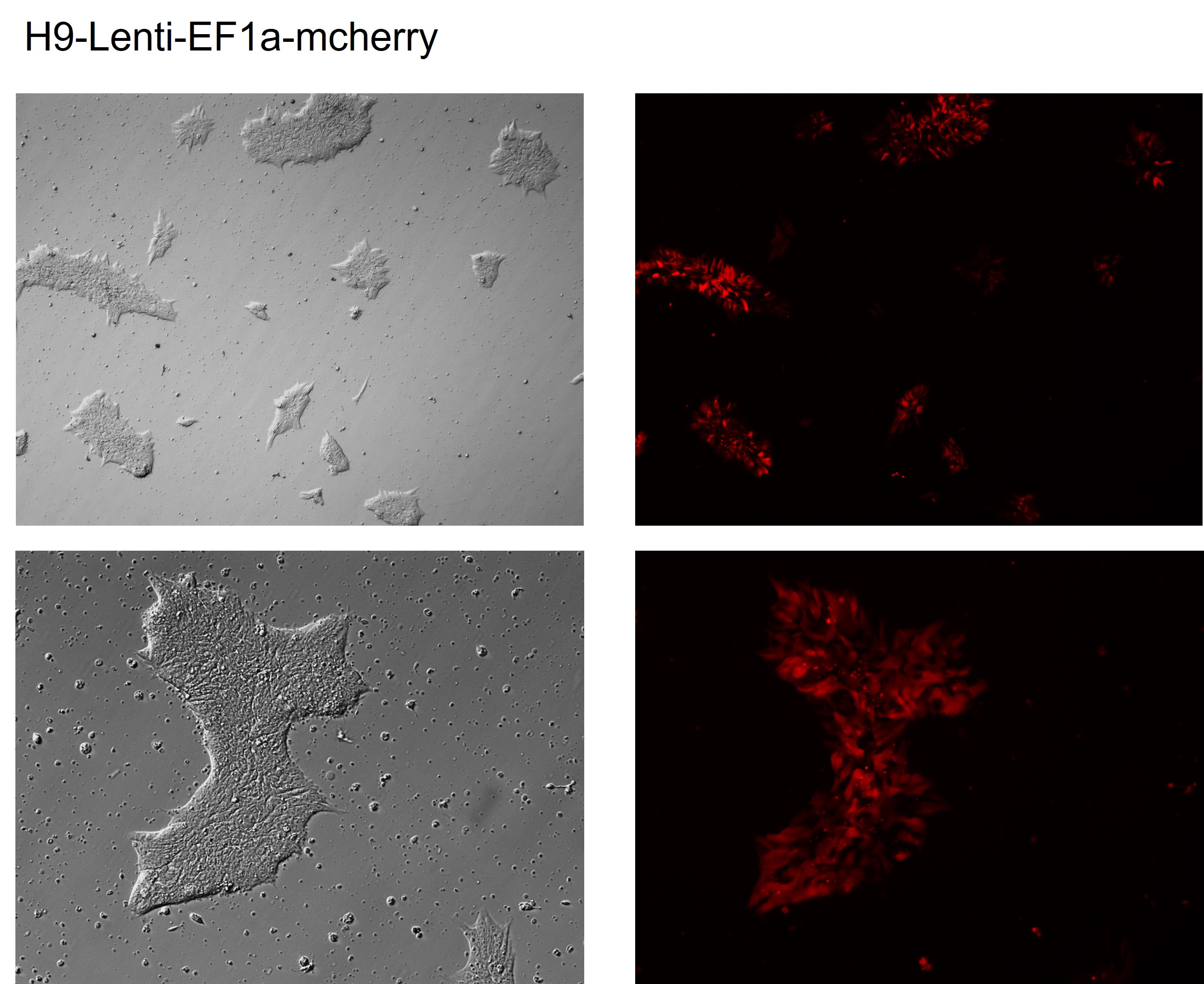
H9::mcherry cyto-reporter line
WAe009-A-1J
General
Cell Line |
|
| hPSCreg name | WAe009-A-1J |
| Cite as: | WAe009-A-1J |
| Alternative name(s) |
H9::mcherry cyto-reporter line
|
| Cell line type | Human embryonic stem cell (hESC) |
| Similar lines |
WAe009-A (WA09, H9) WAe009-A-77 (H9-GFP) WAe009-A-1H (MYL3-KO) WAe009-A-1I (H9::GFP cyto-reporter line) WAe009-A-78 (TBX18-KO) WAe009-A-7 (H9-mHOXA9) WAe009-A-R (H9-NRL-GP, WA09 NRL+/eGFP) WAe009-A-36 (JPH2-KO) WAe009-A-12 (H9_RB1ex3_C7) WAe009-A-13 (H9_RB1ex3_G12LS) WAe009-A-1E (TBX20-KO) WAe009-A-37 (H9-GSX2-tTA:GFP) WAe009-A-99 (dCas9-p300 H9 21) WAe009-A-1V (H9-AAVS1-Teton-KrasG12D) WAe009-A-62 (KCNQ1 KO) WAe009-A-1G (ISL1-KO) WAe009-A-90 (H9_LCCS1) WAe009-A-72 (COL1A2 -/-) WAe009-A-58 (COL4A5 heterozygote) |
| Last update | 26th October 2023 |
| User feedback | |
Provider |
|
| Generator | Memorial Sloan Kettering Cancer Center (MSK) |
External Databases |
|
| BioSamples | SAMEA114549740 |
General Information |
|
| * Is the cell line readily obtainable for third parties? |
Yes Research use: allowed
Clinical use: not allowed
Commercial use: not allowed
|
| Subclone of | |
Donor Information
General Donor Information |
|
| Sex | female |
| Ethnicity | N/A |
Phenotype and Disease related information (Donor) |
|
| Diseases | No disease was diagnosed.
|
| Family history | NO |
| Is the medical history available upon request? | NO |
| Is clinical information available? | NO |
Other Genotyping (Donor) |
|
| Is there genome-wide genotyping or functional data available? |
No
|
Donor Relations |
|
| Other cell lines of this donor | |
External Databases (Donor) |
|
| BioSamples | SAMEA7768918 |
Ethics
Also have a look at the ethics information for the parental line
WAe009-A
.
| Is there an MTA available for the cell line? | No |
| Are you aware of any constraints on the use or distribution of the cell line from the owner or any parties identified in the query above? | No |
hESC Derivation
|
The source cell information can be found in the parental cell line
WAe009-A.
|
Culture Conditions
| Surface coating | Matrigel/Geltrex |
| Feeder cells |
No |
| Passage method |
Enzyme-free cell dissociation
EDTA
|
| Medium |
Essential 8™
|
| Has Rock inhibitor (Y27632) been used at passage previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at cryo previously with this cell line? | Yes |
| Has Rock inhibitor (Y27632) been used at thaw previously with this cell line? | Yes |
Characterisation
Analysis of Undifferentiated Cells
| Marker | Expressed | Immunostaining | RT-PCR | Flow Cytometry | Enzymatic Assay | Expression Profiles |
| POU5F1 (OCT-4) |
Yes |
|||||
| TRA 1-60 |
Yes |
Differentiation Potency
In vitro directed differentiation
Protocol or reference
NC differentiation.jpg
Figure 5. Vagal and sacral NC exhibits distinct behavior in vitro (A) Schematic drawing of experimental design. RFP-tagged hPSC line was used for vagal lineages and GFP-tagged line for sacral lineages. (B) qRT-PCR of SOX10 and HOX genes to confirm the VNC and SNC identity. n = 6 biological replicates (C) Invasion assay of VNC and SNC. Cells crossed the membrane are visualized (left) and quantified using plater reader (right). Scale bars, 50 mm. n = 3 biological replicates. (D) Migration assay of VNC and SNC on PO/LM/FN surface. Cells migrated out of spherical aggregate were imaged (left) and migration distance were quantified (right). Scale bars, 500 mm. n = 3 biological replicates. (E) 3D Matrigel embedded migration assay of co-cultured VNC and SNC. Fluorescence image of cells at 24 (upper) and 96 h (lower), show self-sorting activity. Scale bars, 500 mm. (F) Co-cultured VNC (red) and SNC (green) cells undergoing ENS differentiation with replating at day 10. Scale bars, 100 mm.
Genotyping
Karyotyping (Cell Line) |
|
| Has the cell line karyotype been analysed? |
Yes
|
Other Genotyping (Cell Line) |
|
Genetic Modification
| Genetic modifications not related to a disease |
|


Login to share your feedback, experiences or results with the research community.